Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
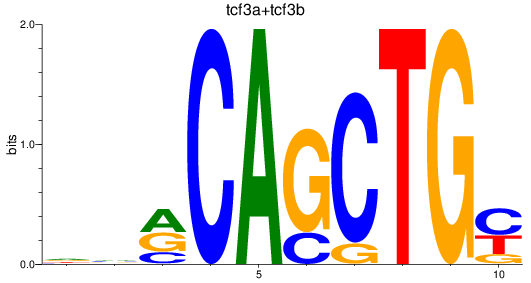
Results for tcf3a+tcf3b
Z-value: 0.99
Transcription factors associated with tcf3a+tcf3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tcf3a
|
ENSDARG00000005915 | transcription factor 3a |
|
tcf3b
|
ENSDARG00000099999 | transcription factor 3b |
|
tcf3b
|
ENSDARG00000112646 | transcription factor 3b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tcf3b | dr11_v1_chr22_-_20342260_20342260 | 0.39 | 1.1e-01 | Click! |
| tcf3a | dr11_v1_chr2_-_57379216_57379216 | -0.10 | 7.1e-01 | Click! |
Activity profile of tcf3a+tcf3b motif
Sorted Z-values of tcf3a+tcf3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_58751504 | 3.80 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr6_+_153146 | 3.20 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr19_+_14109348 | 2.39 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr4_-_20177868 | 2.19 |
ENSDART00000003621
|
sinup
|
siaz-interacting nuclear protein |
| chr21_+_25777425 | 2.18 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr19_-_27564458 | 2.17 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr21_+_5192016 | 2.00 |
ENSDART00000139288
|
si:dkey-121h17.7
|
si:dkey-121h17.7 |
| chr10_-_8046764 | 1.98 |
ENSDART00000099031
|
zgc:136254
|
zgc:136254 |
| chr19_+_7001170 | 1.97 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr13_-_21672131 | 1.87 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr22_-_10541372 | 1.77 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr22_-_10541712 | 1.76 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr1_-_18811517 | 1.72 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr19_-_27570333 | 1.69 |
ENSDART00000146562
ENSDART00000179060 |
si:dkeyp-46h3.5
si:dkeyp-46h3.8
|
si:dkeyp-46h3.5 si:dkeyp-46h3.8 |
| chr1_+_59321629 | 1.69 |
ENSDART00000161981
|
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr19_-_4793263 | 1.68 |
ENSDART00000147510
ENSDART00000141336 ENSDART00000110551 ENSDART00000146684 |
st3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr5_-_68333081 | 1.65 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr2_+_6253246 | 1.65 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr21_-_43666420 | 1.63 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr2_-_17114852 | 1.61 |
ENSDART00000006549
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr16_-_42066523 | 1.59 |
ENSDART00000180538
ENSDART00000058620 |
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr8_-_7232413 | 1.59 |
ENSDART00000092426
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr1_-_34450622 | 1.58 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr12_-_48168135 | 1.54 |
ENSDART00000186624
|
pald1a
|
phosphatase domain containing, paladin 1a |
| chr19_-_27564980 | 1.53 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr22_+_17828267 | 1.51 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr10_+_44699734 | 1.51 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr21_-_19919020 | 1.49 |
ENSDART00000147396
|
ppp1r3b
|
protein phosphatase 1, regulatory subunit 3B |
| chr3_-_27880229 | 1.47 |
ENSDART00000151404
|
abat
|
4-aminobutyrate aminotransferase |
| chr2_-_17115256 | 1.47 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr10_-_25217347 | 1.46 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr3_-_3413669 | 1.44 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr10_-_8053385 | 1.41 |
ENSDART00000142714
|
si:ch211-251f6.7
|
si:ch211-251f6.7 |
| chr10_+_44700103 | 1.37 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr1_+_24387659 | 1.37 |
ENSDART00000130356
|
qdprb2
|
quinoid dihydropteridine reductase b2 |
| chr4_-_13614797 | 1.33 |
ENSDART00000138366
ENSDART00000165212 |
irf5
|
interferon regulatory factor 5 |
| chr7_-_48263516 | 1.31 |
ENSDART00000006619
ENSDART00000142370 ENSDART00000148273 ENSDART00000147968 |
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr11_-_544187 | 1.30 |
ENSDART00000173161
|
CU695207.1
|
|
| chr21_-_30293224 | 1.30 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr11_-_45171139 | 1.28 |
ENSDART00000167036
ENSDART00000161712 ENSDART00000158156 |
syngr2b
|
synaptogyrin 2b |
| chr20_-_20930926 | 1.24 |
ENSDART00000123909
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr2_-_50225411 | 1.23 |
ENSDART00000147117
ENSDART00000000042 |
mcm6l
|
MCM6 minichromosome maintenance deficient 6, like |
| chr23_+_384850 | 1.18 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr6_-_15065376 | 1.17 |
ENSDART00000087797
|
tgfbrap1
|
transforming growth factor, beta receptor associated protein 1 |
| chr8_+_12118097 | 1.17 |
ENSDART00000081819
|
endog
|
endonuclease G |
| chr16_-_17713859 | 1.17 |
ENSDART00000149275
|
zgc:174935
|
zgc:174935 |
| chr9_-_356501 | 1.16 |
ENSDART00000168654
|
zgc:158320
|
zgc:158320 |
| chr24_+_14937205 | 1.15 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr12_-_33357655 | 1.15 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr3_+_40576447 | 1.14 |
ENSDART00000083212
|
fscn1a
|
fascin actin-bundling protein 1a |
| chr8_-_18582922 | 1.13 |
ENSDART00000123917
|
tmem47
|
transmembrane protein 47 |
| chr2_+_30249977 | 1.12 |
ENSDART00000109160
ENSDART00000135171 |
tmem70
|
transmembrane protein 70 |
| chr13_+_36595618 | 1.11 |
ENSDART00000022684
|
cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr15_+_6459847 | 1.11 |
ENSDART00000157250
ENSDART00000065824 |
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr5_+_29851433 | 1.11 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr13_+_11439486 | 1.07 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr8_-_1255321 | 1.07 |
ENSDART00000149605
|
cdc14b
|
cell division cycle 14B |
| chr1_-_156375 | 1.06 |
ENSDART00000160221
|
pcid2
|
PCI domain containing 2 |
| chr14_-_16810401 | 1.05 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr20_+_400487 | 1.05 |
ENSDART00000108872
|
ndufaf4
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 4 |
| chr10_+_1052591 | 1.03 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr23_-_31647793 | 1.00 |
ENSDART00000145621
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr20_+_48116476 | 0.99 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr8_+_144154 | 0.98 |
ENSDART00000164099
|
snx2
|
sorting nexin 2 |
| chr19_+_40069524 | 0.98 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr9_+_28232522 | 0.97 |
ENSDART00000031761
|
fzd5
|
frizzled class receptor 5 |
| chr5_-_53004052 | 0.97 |
ENSDART00000157978
|
mrps30
|
mitochondrial ribosomal protein S30 |
| chr21_+_5974590 | 0.96 |
ENSDART00000098499
|
dck
|
deoxycytidine kinase |
| chr20_-_164300 | 0.96 |
ENSDART00000183354
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr5_-_69948099 | 0.96 |
ENSDART00000034639
ENSDART00000191111 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr14_-_24109580 | 0.96 |
ENSDART00000181415
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr13_-_9442942 | 0.95 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr23_-_16692312 | 0.95 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr2_+_8261109 | 0.95 |
ENSDART00000024677
|
dia1a
|
deleted in autism 1a |
| chr12_+_20693743 | 0.94 |
ENSDART00000153023
ENSDART00000153370 |
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr7_-_18881358 | 0.94 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr18_+_45573416 | 0.93 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr24_-_32522587 | 0.93 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr14_-_899170 | 0.93 |
ENSDART00000165211
ENSDART00000031992 |
rgs14a
|
regulator of G protein signaling 14a |
| chr13_+_2894536 | 0.93 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr6_-_1820606 | 0.93 |
ENSDART00000183228
|
FO834857.1
|
|
| chr7_-_19364813 | 0.92 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr3_+_40164129 | 0.92 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr12_+_20691310 | 0.92 |
ENSDART00000064335
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr16_+_27614989 | 0.91 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr14_+_21828993 | 0.91 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr5_-_63644938 | 0.91 |
ENSDART00000050865
|
surf4l
|
surfeit gene 4, like |
| chr5_-_69944084 | 0.90 |
ENSDART00000188557
ENSDART00000127782 |
ugt2a4
|
UDP glucuronosyltransferase 2 family, polypeptide A4 |
| chr5_-_12743196 | 0.90 |
ENSDART00000188976
ENSDART00000137705 |
lztr1
|
leucine-zipper-like transcription regulator 1 |
| chr4_-_4834347 | 0.90 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr13_+_33462232 | 0.90 |
ENSDART00000177841
|
zgc:136302
|
zgc:136302 |
| chr22_-_17677947 | 0.88 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr13_-_31938512 | 0.88 |
ENSDART00000026726
ENSDART00000182666 |
diexf
|
digestive organ expansion factor homolog |
| chr12_-_10512911 | 0.88 |
ENSDART00000124562
ENSDART00000106163 |
zgc:152977
|
zgc:152977 |
| chr16_+_39159752 | 0.87 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr15_+_43906043 | 0.87 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr20_+_1316495 | 0.87 |
ENSDART00000064439
|
nup43
|
nucleoporin 43 |
| chr5_+_62052538 | 0.87 |
ENSDART00000141574
|
si:dkey-35m8.1
|
si:dkey-35m8.1 |
| chr1_-_47114310 | 0.87 |
ENSDART00000144899
ENSDART00000053157 |
setd4
|
SET domain containing 4 |
| chr7_-_19369002 | 0.87 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr18_+_5454341 | 0.87 |
ENSDART00000192649
|
dtwd1
|
DTW domain containing 1 |
| chr5_+_26212621 | 0.86 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr20_-_20932760 | 0.86 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr16_+_54641230 | 0.85 |
ENSDART00000157641
ENSDART00000159540 |
fbxo43
|
F-box protein 43 |
| chr8_+_36500308 | 0.85 |
ENSDART00000098701
|
slc7a4
|
solute carrier family 7, member 4 |
| chr23_-_18057270 | 0.84 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr11_+_13041341 | 0.84 |
ENSDART00000166126
ENSDART00000162074 ENSDART00000170889 |
btf3l4
|
basic transcription factor 3-like 4 |
| chr18_+_45573251 | 0.84 |
ENSDART00000191309
|
kifc3
|
kinesin family member C3 |
| chr6_+_59991076 | 0.83 |
ENSDART00000163575
|
CABZ01100888.1
|
|
| chr13_-_39254 | 0.82 |
ENSDART00000093222
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr6_+_10338554 | 0.82 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr17_+_16090436 | 0.82 |
ENSDART00000136059
ENSDART00000138734 |
znf395a
|
zinc finger protein 395a |
| chr20_+_1196386 | 0.82 |
ENSDART00000041192
|
lyrm2
|
LYR motif containing 2 |
| chr14_+_591216 | 0.81 |
ENSDART00000169624
|
zgc:158257
|
zgc:158257 |
| chr8_+_36500061 | 0.81 |
ENSDART00000185840
|
slc7a4
|
solute carrier family 7, member 4 |
| chr23_+_26079467 | 0.81 |
ENSDART00000129617
|
atp6ap1b
|
ATPase H+ transporting accessory protein 1b |
| chr17_+_4368859 | 0.81 |
ENSDART00000055385
|
crls1
|
cardiolipin synthase 1 |
| chr5_+_37744625 | 0.80 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr14_-_34605607 | 0.80 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr13_-_8692860 | 0.80 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr14_-_25930182 | 0.80 |
ENSDART00000018651
ENSDART00000147991 |
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr12_-_25380028 | 0.80 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr14_-_16754262 | 0.80 |
ENSDART00000001159
|
mgat4b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B |
| chr3_+_25990052 | 0.79 |
ENSDART00000007398
|
gcat
|
glycine C-acetyltransferase |
| chr10_+_10387328 | 0.79 |
ENSDART00000080904
|
sardh
|
sarcosine dehydrogenase |
| chr23_+_17522867 | 0.78 |
ENSDART00000002714
|
slc17a9b
|
solute carrier family 17 (vesicular nucleotide transporter), member 9b |
| chr11_-_34219211 | 0.78 |
ENSDART00000098472
|
tmem44
|
transmembrane protein 44 |
| chr22_-_5171829 | 0.78 |
ENSDART00000140313
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr5_-_57624425 | 0.77 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr1_+_12302073 | 0.77 |
ENSDART00000164045
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr6_-_14038804 | 0.77 |
ENSDART00000184606
ENSDART00000184609 |
etv5b
|
ets variant 5b |
| chr5_-_11573490 | 0.77 |
ENSDART00000109577
|
FO704871.1
|
|
| chr14_-_33278084 | 0.76 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr5_+_34549845 | 0.76 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr19_+_15441022 | 0.76 |
ENSDART00000098970
ENSDART00000140276 |
lin28a
|
lin-28 homolog A (C. elegans) |
| chr14_-_25928899 | 0.75 |
ENSDART00000143518
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr14_-_33277743 | 0.75 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr2_+_11031360 | 0.75 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr17_+_1360192 | 0.74 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr15_+_1199407 | 0.74 |
ENSDART00000163827
|
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr19_-_425145 | 0.74 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr10_+_2841205 | 0.74 |
ENSDART00000131505
ENSDART00000055869 |
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr3_-_40275096 | 0.73 |
ENSDART00000141578
|
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr10_-_14929630 | 0.73 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr14_-_24110062 | 0.72 |
ENSDART00000177062
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr21_-_44772738 | 0.72 |
ENSDART00000026178
|
kif4
|
kinesin family member 4 |
| chr16_+_25259313 | 0.71 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr8_-_13735572 | 0.71 |
ENSDART00000139642
|
si:dkey-258f14.7
|
si:dkey-258f14.7 |
| chr1_-_34450784 | 0.71 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr1_-_524433 | 0.70 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr5_+_1515938 | 0.70 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr17_-_51818659 | 0.70 |
ENSDART00000111389
ENSDART00000157244 |
exd2
|
exonuclease 3'-5' domain containing 2 |
| chr1_+_30946231 | 0.69 |
ENSDART00000022841
|
metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr12_+_24060894 | 0.68 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr20_-_23254876 | 0.68 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr12_-_10505986 | 0.68 |
ENSDART00000152672
|
zgc:152977
|
zgc:152977 |
| chr9_+_34148714 | 0.68 |
ENSDART00000078051
|
gpr161
|
G protein-coupled receptor 161 |
| chr3_+_60607241 | 0.67 |
ENSDART00000167512
|
mfsd11
|
major facilitator superfamily domain containing 11 |
| chr21_+_13387965 | 0.67 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr22_-_6420239 | 0.67 |
ENSDART00000148385
|
zgc:171699
|
zgc:171699 |
| chr20_+_14968031 | 0.67 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr5_-_65121747 | 0.66 |
ENSDART00000165556
|
tor2a
|
torsin family 2, member A |
| chr19_+_15485556 | 0.66 |
ENSDART00000079014
|
pdik1l
|
PDLIM1 interacting kinase 1 like |
| chr16_+_813780 | 0.66 |
ENSDART00000162474
ENSDART00000161774 |
irx1a
|
iroquois homeobox 1a |
| chr19_-_48010490 | 0.66 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr1_-_44048798 | 0.66 |
ENSDART00000073746
|
si:ch73-109d9.2
|
si:ch73-109d9.2 |
| chr18_+_30441740 | 0.66 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr14_+_30340251 | 0.66 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr25_+_32390794 | 0.66 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr5_-_23696926 | 0.65 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr12_+_3262564 | 0.65 |
ENSDART00000184264
|
tmem101
|
transmembrane protein 101 |
| chr15_-_25099679 | 0.65 |
ENSDART00000154628
|
rflnb
|
refilin B |
| chr14_+_38786298 | 0.65 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr11_+_24313931 | 0.64 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr8_-_34427364 | 0.64 |
ENSDART00000112854
ENSDART00000161282 ENSDART00000113230 |
gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr19_+_42660158 | 0.64 |
ENSDART00000018328
|
fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chr19_+_14454306 | 0.63 |
ENSDART00000161965
|
zdhhc18b
|
zinc finger, DHHC-type containing 18b |
| chr6_+_38896158 | 0.63 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
| chr5_+_36781732 | 0.63 |
ENSDART00000087191
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr19_-_30810328 | 0.63 |
ENSDART00000184875
|
myclb
|
MYCL proto-oncogene, bHLH transcription factor b |
| chr15_-_704408 | 0.63 |
ENSDART00000156200
ENSDART00000166404 ENSDART00000131040 |
zgc:174574
|
zgc:174574 |
| chr17_-_24884966 | 0.63 |
ENSDART00000136142
|
gale
|
UDP-galactose-4-epimerase |
| chr19_+_28187480 | 0.62 |
ENSDART00000183825
|
irx4b
|
iroquois homeobox 4b |
| chr23_-_31648026 | 0.62 |
ENSDART00000133569
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr24_-_11905911 | 0.61 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr10_+_17345 | 0.61 |
ENSDART00000180325
|
tiprl
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr1_+_13930625 | 0.61 |
ENSDART00000111026
|
noctb
|
nocturnin b |
| chr21_+_28747069 | 0.61 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr25_+_3294150 | 0.61 |
ENSDART00000030683
|
tmpob
|
thymopoietin b |
| chr6_-_11759860 | 0.61 |
ENSDART00000151296
|
si:ch211-162i14.1
|
si:ch211-162i14.1 |
| chr11_-_44898129 | 0.61 |
ENSDART00000157615
|
eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr21_-_34261677 | 0.60 |
ENSDART00000124649
ENSDART00000172381 ENSDART00000064320 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr5_+_28271412 | 0.60 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr19_-_12212692 | 0.60 |
ENSDART00000142077
ENSDART00000151599 ENSDART00000140834 ENSDART00000078781 |
znf706
|
zinc finger protein 706 |
| chr1_+_12301913 | 0.60 |
ENSDART00000165733
|
gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr5_-_13848296 | 0.60 |
ENSDART00000127109
|
figla
|
folliculogenesis specific bHLH transcription factor |
| chr16_-_8132742 | 0.60 |
ENSDART00000104323
|
snrka
|
SNF related kinase a |
| chr23_+_39346774 | 0.60 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr23_+_43718865 | 0.60 |
ENSDART00000175192
|
anapc10
|
anaphase promoting complex subunit 10 |
| chr2_-_49854432 | 0.59 |
ENSDART00000180731
|
blvra
|
biliverdin reductase A |
Network of associatons between targets according to the STRING database.
First level regulatory network of tcf3a+tcf3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.6 | 0.6 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.5 | 1.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.5 | 1.4 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.4 | 2.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.4 | 1.1 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.3 | 1.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.3 | 1.5 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.3 | 1.4 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.3 | 1.3 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.3 | 3.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.3 | 0.8 | GO:0097676 | histone H3-K36 dimethylation(GO:0097676) |
| 0.2 | 1.7 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.2 | 0.7 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.2 | 1.7 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 0.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 1.1 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.2 | 0.7 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 0.2 | 0.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 0.7 | GO:1903011 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.2 | 3.2 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 1.3 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.2 | 0.7 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.2 | 0.5 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 0.8 | GO:0048209 | regulation of vesicle targeting, to, from or within Golgi(GO:0048209) |
| 0.2 | 0.8 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.2 | 0.5 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 0.5 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.2 | 2.2 | GO:0001840 | neural plate development(GO:0001840) |
| 0.2 | 0.5 | GO:0072387 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.2 | 1.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.1 | GO:2000405 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.1 | 0.6 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 1.5 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.6 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.7 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.7 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.1 | 2.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.1 | 0.4 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.7 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 1.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 1.0 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 2.4 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.2 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.6 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.8 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 0.9 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.8 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.9 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 1.1 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 0.9 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.1 | 0.3 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.4 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.1 | 0.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.8 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.7 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.0 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.6 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.1 | 0.5 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 0.3 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.5 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.1 | 1.6 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.6 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.1 | 1.0 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.8 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.4 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 0.7 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.1 | 0.4 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.2 | GO:0070861 | regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.2 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 | 0.3 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.4 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.1 | 0.3 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 1.1 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.1 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 0.5 | GO:0043931 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.1 | 1.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.4 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.4 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 0.4 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.1 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 0.7 | GO:0008585 | female gonad development(GO:0008585) |
| 0.1 | 1.4 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 0.5 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.3 | GO:0034163 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 1.1 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 0.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.6 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 3.1 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 0.5 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 0.8 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.1 | 0.6 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.1 | 0.2 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.9 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 0.8 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.1 | 0.3 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.1 | 2.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.9 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.1 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 1.0 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.4 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.2 | GO:0007620 | mating(GO:0007618) copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.4 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.7 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.4 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.3 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.3 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.3 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.0 | GO:0014014 | negative regulation of gliogenesis(GO:0014014) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.0 | 0.1 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.0 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.0 | 0.4 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0032447 | protein urmylation(GO:0032447) |
| 0.0 | 0.4 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.7 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.9 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.3 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.6 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0046824 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.2 | GO:0035307 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 2.6 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 1.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.4 | GO:1904103 | regulation of convergent extension involved in gastrulation(GO:1904103) |
| 0.0 | 1.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.5 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 3.5 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.3 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.2 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0010459 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.7 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.9 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0090234 | regulation of centromere complex assembly(GO:0090230) regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.0 | 0.4 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.3 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 1.4 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 2.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.5 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.9 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.4 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 5.2 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.1 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.6 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.3 | GO:0048799 | organ maturation(GO:0048799) |
| 0.0 | 0.7 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.7 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0048794 | swim bladder development(GO:0048794) |
| 0.0 | 0.4 | GO:0009225 | nucleotide-sugar metabolic process(GO:0009225) |
| 0.0 | 0.7 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.9 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.5 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.2 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 0.5 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.3 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0042478 | regulation of eye photoreceptor cell development(GO:0042478) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0019827 | stem cell population maintenance(GO:0019827) |
| 0.0 | 0.3 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.1 | GO:1901021 | positive regulation of calcium ion transmembrane transporter activity(GO:1901021) positive regulation of voltage-gated calcium channel activity(GO:1901387) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.2 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 1.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.6 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.7 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.1 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.2 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 1.0 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.2 | GO:1903307 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) positive regulation of regulated secretory pathway(GO:1903307) |
| 0.0 | 0.6 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.2 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.5 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.2 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.5 | GO:0017015 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.0 | 0.3 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.9 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.2 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.3 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 1.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.0 | GO:0060254 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.0 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte differentiation(GO:0043363) nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0008344 | adult locomotory behavior(GO:0008344) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.3 | GO:0046847 | filopodium assembly(GO:0046847) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.3 | 1.4 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.4 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.8 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.1 | 0.7 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 1.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 0.4 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.8 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 0.6 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.5 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 0.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.4 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 1.5 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.4 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.8 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.2 | GO:0072380 | TRC complex(GO:0072380) |
| 0.1 | 0.9 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 3.0 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 0.2 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.1 | 1.4 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.5 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.4 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 3.1 | GO:0005657 | replication fork(GO:0005657) |
| 0.1 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.4 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.5 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.5 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.3 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 3.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.4 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.8 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.3 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 1.3 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.2 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 3.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.4 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.7 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.1 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 2.2 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.6 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 1.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 5.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.4 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 5.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.0 | GO:0098573 | intrinsic component of mitochondrial membrane(GO:0098573) |
| 0.0 | 0.8 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 1.0 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.6 | 3.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.6 | 2.9 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.5 | 1.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.5 | 1.4 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.4 | 1.5 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.3 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.3 | 1.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 1.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.2 | 0.7 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.2 | 1.7 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.2 | 0.6 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 0.8 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.2 | 0.8 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 1.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 3.2 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 0.7 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.2 | 0.5 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.2 | 2.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 1.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.3 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.1 | 1.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 0.5 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 1.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.4 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.1 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:1990715 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 2.4 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.8 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.4 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0004649 | poly(ADP-ribose) glycohydrolase activity(GO:0004649) |
| 0.1 | 0.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.1 | 1.5 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.3 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.4 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 1.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.1 | 0.4 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.1 | 2.8 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.3 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.1 | 1.5 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 1.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.4 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 1.1 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 0.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.5 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.5 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.1 | 1.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0004776 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.1 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 2.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.1 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.1 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 4.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.8 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 1.9 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.6 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 1.3 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.3 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.9 | GO:0017136 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) |
| 0.0 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.3 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.7 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.2 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.7 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 1.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.7 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.8 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.3 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 2.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 1.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.9 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.0 | 0.6 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 5.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.4 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 1.1 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 0.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 1.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 1.0 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 2.8 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 1.1 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.3 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.1 | 1.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 5.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.3 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 2.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 2.2 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 0.2 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.1 | 1.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.9 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.8 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 1.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.2 | 1.8 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.4 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 1.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 1.4 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 3.9 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 3.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.3 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 2.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.6 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 2.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.5 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.7 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.9 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.1 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.0 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |