Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
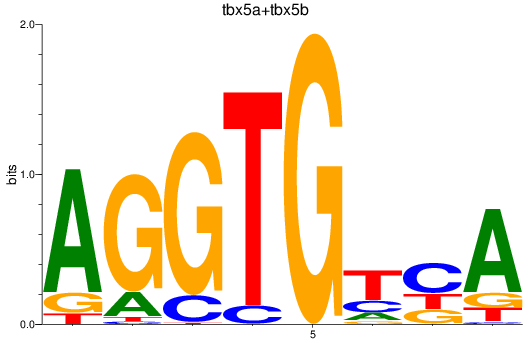
Results for tbx5a+tbx5b
Z-value: 1.28
Transcription factors associated with tbx5a+tbx5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx5a
|
ENSDARG00000024894 | T-box transcription factor 5a |
|
tbx5b
|
ENSDARG00000092060 | T-box transcription factor 5b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx5a | dr11_v1_chr5_-_72289648_72289648 | 0.76 | 2.7e-04 | Click! |
| tbx5b | dr11_v1_chr5_+_23152282_23152282 | -0.34 | 1.7e-01 | Click! |
Activity profile of tbx5a+tbx5b motif
Sorted Z-values of tbx5a+tbx5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_-_10021341 | 4.69 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr24_-_10006158 | 4.61 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr3_+_43086548 | 4.53 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr24_-_9997948 | 4.51 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_+_10027902 | 4.23 |
ENSDART00000175961
ENSDART00000172773 |
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr11_+_29537756 | 4.18 |
ENSDART00000103388
|
wu:fi42e03
|
wu:fi42e03 |
| chr20_-_43743700 | 4.03 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr10_+_19596214 | 3.74 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr19_-_34999379 | 3.57 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr10_+_19569052 | 3.56 |
ENSDART00000058425
|
CABZ01059627.1
|
|
| chr10_+_19595009 | 3.19 |
ENSDART00000112276
|
zgc:173837
|
zgc:173837 |
| chr11_-_6452444 | 3.10 |
ENSDART00000137879
ENSDART00000134957 ENSDART00000004483 |
larp6b
|
La ribonucleoprotein domain family, member 6b |
| chr4_-_77114795 | 2.73 |
ENSDART00000144849
|
CU467646.2
|
|
| chr10_-_21542702 | 2.40 |
ENSDART00000146761
ENSDART00000134502 |
zgc:165539
|
zgc:165539 |
| chr5_-_68333081 | 2.17 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr20_-_14114078 | 1.82 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr21_+_30010061 | 1.66 |
ENSDART00000182163
|
ttc1
|
tetratricopeptide repeat domain 1 |
| chr22_+_39096911 | 1.66 |
ENSDART00000157127
ENSDART00000153841 |
lmcd1
|
LIM and cysteine-rich domains 1 |
| chr19_-_34995629 | 1.65 |
ENSDART00000141704
|
si:rp71-45k5.2
|
si:rp71-45k5.2 |
| chr13_+_2894536 | 1.60 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr23_-_31266586 | 1.54 |
ENSDART00000139746
|
si:dkey-261l7.2
|
si:dkey-261l7.2 |
| chr3_+_49074008 | 1.53 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr21_-_35325466 | 1.49 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr20_+_9124369 | 1.45 |
ENSDART00000064150
|
si:ch211-59d15.9
|
si:ch211-59d15.9 |
| chr24_-_38079261 | 1.43 |
ENSDART00000105662
|
crp1
|
C-reactive protein 1 |
| chr4_+_9478500 | 1.42 |
ENSDART00000030738
|
lmf2b
|
lipase maturation factor 2b |
| chr18_+_16053455 | 1.39 |
ENSDART00000189163
ENSDART00000188269 |
FO834850.1
|
|
| chr3_+_32099507 | 1.34 |
ENSDART00000044238
|
zgc:92066
|
zgc:92066 |
| chr4_+_14981854 | 1.33 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr8_-_38317914 | 1.30 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr22_+_26853254 | 1.30 |
ENSDART00000182487
|
tmem186
|
transmembrane protein 186 |
| chr10_-_2522588 | 1.28 |
ENSDART00000081926
|
CU856539.1
|
|
| chr19_+_791538 | 1.27 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr3_+_51563695 | 1.26 |
ENSDART00000008607
|
ttyh2l
|
tweety homolog 2, like |
| chr19_-_8812891 | 1.26 |
ENSDART00000151134
ENSDART00000025385 ENSDART00000180291 |
cers2a
|
ceramide synthase 2a |
| chr21_+_39197628 | 1.25 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr5_+_26213874 | 1.25 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr7_-_38689562 | 1.21 |
ENSDART00000167209
|
aplnr2
|
apelin receptor 2 |
| chr13_-_51846224 | 1.20 |
ENSDART00000184663
|
LT631684.2
|
|
| chr22_-_17652914 | 1.19 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr21_-_34972872 | 1.19 |
ENSDART00000023838
|
lipia
|
lipase, member Ia |
| chr23_+_44614056 | 1.17 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr5_-_32338866 | 1.16 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr15_-_25365319 | 1.15 |
ENSDART00000152651
|
cluha
|
clustered mitochondria (cluA/CLU1) homolog a |
| chr15_+_20530649 | 1.14 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr5_+_57658898 | 1.13 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr19_+_37135700 | 1.12 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr9_+_27720428 | 1.12 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr18_-_14274803 | 1.10 |
ENSDART00000166643
|
mlycd
|
malonyl-CoA decarboxylase |
| chr24_-_11908115 | 1.08 |
ENSDART00000184329
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr22_-_17653143 | 1.07 |
ENSDART00000089171
|
hmha1b
|
histocompatibility (minor) HA-1 b |
| chr14_+_41318881 | 1.05 |
ENSDART00000192137
|
xkrx
|
XK, Kell blood group complex subunit-related, X-linked |
| chr3_-_54607166 | 1.04 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr3_+_58472305 | 1.04 |
ENSDART00000154122
|
si:ch211-165g14.1
|
si:ch211-165g14.1 |
| chr15_-_45538773 | 1.03 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr7_-_55648336 | 1.03 |
ENSDART00000147792
ENSDART00000135304 ENSDART00000131923 |
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr1_+_19602389 | 1.01 |
ENSDART00000088933
ENSDART00000141579 ENSDART00000111555 |
fbxo10
|
F-box protein 10 |
| chr15_+_43906043 | 1.01 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr24_+_26337623 | 1.01 |
ENSDART00000145637
|
mynn
|
myoneurin |
| chr2_-_38114370 | 1.00 |
ENSDART00000131837
|
chd8
|
chromodomain helicase DNA binding protein 8 |
| chr3_+_24062748 | 0.98 |
ENSDART00000188574
|
cbx1a
|
chromobox homolog 1a (HP1 beta homolog Drosophila) |
| chr14_+_48862987 | 0.97 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr1_+_49814461 | 0.96 |
ENSDART00000132405
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr21_+_39197418 | 0.96 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr20_-_38778479 | 0.95 |
ENSDART00000185599
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr5_-_64355227 | 0.94 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr3_+_40255408 | 0.94 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr12_+_33460794 | 0.93 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr6_-_39199070 | 0.93 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr2_-_23391266 | 0.91 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr1_-_524433 | 0.90 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr8_-_39984593 | 0.87 |
ENSDART00000140127
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr2_+_11028923 | 0.86 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr10_+_38610741 | 0.86 |
ENSDART00000126444
|
mmp13a
|
matrix metallopeptidase 13a |
| chr5_+_37966505 | 0.86 |
ENSDART00000127648
|
pafah1b2
|
platelet-activating factor acetylhydrolase 1b, catalytic subunit 2 |
| chr20_+_1316803 | 0.85 |
ENSDART00000152586
ENSDART00000152165 |
nup43
|
nucleoporin 43 |
| chr10_-_1961930 | 0.85 |
ENSDART00000122446
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr8_-_3312384 | 0.84 |
ENSDART00000035965
|
fut9b
|
fucosyltransferase 9b |
| chr10_+_1052591 | 0.82 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr22_+_24603930 | 0.81 |
ENSDART00000180240
ENSDART00000164256 |
CU655842.1
|
|
| chr2_-_9607879 | 0.81 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr10_+_34001444 | 0.80 |
ENSDART00000149934
|
kl
|
klotho |
| chr22_+_26600834 | 0.80 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr8_-_38201415 | 0.80 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr7_+_48297842 | 0.79 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr15_-_19443997 | 0.78 |
ENSDART00000114936
|
esamb
|
endothelial cell adhesion molecule b |
| chr7_+_40081630 | 0.77 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr5_-_69482891 | 0.77 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr8_-_39978767 | 0.77 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr3_-_60027255 | 0.76 |
ENSDART00000189252
ENSDART00000154684 |
recql5
|
RecQ helicase-like 5 |
| chr16_+_10264601 | 0.76 |
ENSDART00000186167
|
mrs2
|
MRS2 magnesium transporter |
| chr5_+_29652513 | 0.76 |
ENSDART00000035400
|
tsc1a
|
TSC complex subunit 1a |
| chr8_+_3431671 | 0.75 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr4_-_18512550 | 0.75 |
ENSDART00000045639
|
rint1
|
RAD50 interactor 1 |
| chr6_-_37499574 | 0.74 |
ENSDART00000023163
|
jade3
|
jade family PHD finger 3 |
| chr11_-_18015534 | 0.74 |
ENSDART00000181953
|
qrich1
|
glutamine-rich 1 |
| chr10_+_16501699 | 0.73 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr23_-_12158685 | 0.73 |
ENSDART00000135035
|
fam217b
|
family with sequence similarity 217, member B |
| chr15_-_11681653 | 0.73 |
ENSDART00000180160
|
fkrp
|
fukutin related protein |
| chr12_-_36045283 | 0.73 |
ENSDART00000160646
|
gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr17_-_15029639 | 0.72 |
ENSDART00000142941
|
ero1a
|
endoplasmic reticulum oxidoreductase alpha |
| chr25_-_29363934 | 0.72 |
ENSDART00000166889
|
nptna
|
neuroplastin a |
| chr21_-_36396334 | 0.71 |
ENSDART00000183627
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr12_-_3053873 | 0.70 |
ENSDART00000023796
ENSDART00000137148 |
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr8_-_1838315 | 0.70 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr25_+_11281970 | 0.70 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr17_+_49276061 | 0.69 |
ENSDART00000059255
|
zgc:113176
|
zgc:113176 |
| chr12_+_1398404 | 0.69 |
ENSDART00000026303
|
rasd1
|
RAS, dexamethasone-induced 1 |
| chr18_+_36664654 | 0.69 |
ENSDART00000128707
ENSDART00000098972 |
capn12
|
calpain 12 |
| chr24_-_32173754 | 0.69 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr11_-_41220794 | 0.69 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr4_+_20486041 | 0.69 |
ENSDART00000017572
|
ints13
|
integrator complex subunit 13 |
| chr6_-_11759860 | 0.68 |
ENSDART00000151296
|
si:ch211-162i14.1
|
si:ch211-162i14.1 |
| chr2_-_23390779 | 0.68 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr9_+_25840720 | 0.67 |
ENSDART00000024572
|
gtdc1
|
glycosyltransferase-like domain containing 1 |
| chr9_-_4606463 | 0.66 |
ENSDART00000179110
|
galnt13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 |
| chr10_+_13000669 | 0.66 |
ENSDART00000158919
ENSDART00000172625 |
lpar1
|
lysophosphatidic acid receptor 1 |
| chr25_-_6049339 | 0.65 |
ENSDART00000075184
|
snx1a
|
sorting nexin 1a |
| chr6_+_13232934 | 0.64 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr6_+_54498220 | 0.64 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr5_+_6670945 | 0.64 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr16_+_10264286 | 0.63 |
ENSDART00000091377
|
mrs2
|
MRS2 magnesium transporter |
| chr1_+_49415281 | 0.63 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr17_-_51262430 | 0.63 |
ENSDART00000163222
|
trappc12
|
trafficking protein particle complex 12 |
| chr10_-_45029041 | 0.63 |
ENSDART00000167878
|
polm
|
polymerase (DNA directed), mu |
| chr5_-_26893310 | 0.62 |
ENSDART00000126669
|
lman2lb
|
lectin, mannose-binding 2-like b |
| chr13_-_33246154 | 0.62 |
ENSDART00000130127
|
rtf1
|
RTF1 homolog, Paf1/RNA polymerase II complex component |
| chr21_-_2287589 | 0.62 |
ENSDART00000161554
|
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr9_+_34221997 | 0.62 |
ENSDART00000028617
|
mpc2
|
mitochondrial pyruvate carrier 2 |
| chr3_+_15773991 | 0.61 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr7_+_15313443 | 0.61 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr7_-_69795488 | 0.60 |
ENSDART00000162414
|
USP53 (1 of many)
|
ubiquitin specific peptidase 53 |
| chr13_-_7031033 | 0.60 |
ENSDART00000193211
|
CABZ01061524.1
|
|
| chr3_+_34986837 | 0.60 |
ENSDART00000190341
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr3_+_26288981 | 0.60 |
ENSDART00000163500
|
rhot1a
|
ras homolog family member T1a |
| chr25_+_31405266 | 0.60 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr21_-_14251306 | 0.60 |
ENSDART00000114715
ENSDART00000181380 |
man1b1a
|
mannosidase, alpha, class 1B, member 1a |
| chr15_-_1765098 | 0.59 |
ENSDART00000149980
ENSDART00000093074 |
bud23
|
BUD23, rRNA methyltransferase and ribosome maturation factor |
| chr15_-_1622468 | 0.59 |
ENSDART00000149008
ENSDART00000034456 |
kpna4
|
karyopherin alpha 4 (importin alpha 3) |
| chr3_+_35542067 | 0.59 |
ENSDART00000146529
ENSDART00000084549 |
rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr5_-_31856681 | 0.59 |
ENSDART00000187817
|
pkn3
|
protein kinase N3 |
| chr6_-_25165693 | 0.58 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr3_-_23604396 | 0.57 |
ENSDART00000078423
|
atp5mc1
|
ATP synthase membrane subunit c locus 1 |
| chr18_+_45666489 | 0.57 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr1_+_41131481 | 0.57 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr7_-_54320088 | 0.56 |
ENSDART00000172396
|
fadd
|
Fas (tnfrsf6)-associated via death domain |
| chr2_-_32237916 | 0.56 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr23_-_6982966 | 0.56 |
ENSDART00000033774
ENSDART00000169965 |
edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr8_-_10947205 | 0.55 |
ENSDART00000164467
|
pqlc2
|
PQ loop repeat containing 2 |
| chr15_+_40079468 | 0.55 |
ENSDART00000154947
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr10_+_8550435 | 0.54 |
ENSDART00000185664
|
tbc1d10ab
|
TBC1 domain family, member 10Ab |
| chr25_-_6292270 | 0.54 |
ENSDART00000130809
|
wdr61
|
WD repeat domain 61 |
| chr1_+_45002971 | 0.54 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr20_+_3339248 | 0.54 |
ENSDART00000108955
|
mfsd4b
|
major facilitator superfamily domain containing 4B |
| chr2_-_15041846 | 0.54 |
ENSDART00000139050
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr9_-_28255029 | 0.54 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr21_-_38618540 | 0.54 |
ENSDART00000036600
|
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr25_-_6292560 | 0.53 |
ENSDART00000153496
|
wdr61
|
WD repeat domain 61 |
| chr24_-_2312868 | 0.53 |
ENSDART00000140125
ENSDART00000138432 |
cul2
|
cullin 2 |
| chr8_+_32722842 | 0.53 |
ENSDART00000147594
|
hmcn2
|
hemicentin 2 |
| chr13_+_2538636 | 0.53 |
ENSDART00000168927
|
plpp4
|
phospholipid phosphatase 4 |
| chr9_-_27720612 | 0.52 |
ENSDART00000000566
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr2_-_58257624 | 0.52 |
ENSDART00000098940
|
foxl2b
|
forkhead box L2b |
| chr13_-_31346392 | 0.52 |
ENSDART00000134343
|
rrp12
|
ribosomal RNA processing 12 homolog |
| chr13_+_40334727 | 0.51 |
ENSDART00000074950
|
slc25a28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr15_-_28587490 | 0.51 |
ENSDART00000186196
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr16_+_26439518 | 0.51 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr2_+_38226624 | 0.51 |
ENSDART00000131384
|
si:ch211-14a17.11
|
si:ch211-14a17.11 |
| chr2_-_58075414 | 0.51 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr10_+_42521943 | 0.50 |
ENSDART00000010420
ENSDART00000075303 |
actr1
|
ARP1 actin related protein 1, centractin |
| chr7_+_22718251 | 0.50 |
ENSDART00000027718
ENSDART00000143341 |
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr14_-_7245971 | 0.50 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr6_-_10725847 | 0.49 |
ENSDART00000184567
|
sp3b
|
Sp3b transcription factor |
| chr21_+_6136981 | 0.49 |
ENSDART00000065859
ENSDART00000166287 |
cdk9
|
cyclin-dependent kinase 9 (CDC2-related kinase) |
| chr16_+_50434668 | 0.49 |
ENSDART00000193500
|
IGLON5
|
zgc:110372 |
| chr21_-_14826066 | 0.49 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr23_-_27505825 | 0.49 |
ENSDART00000137229
ENSDART00000013797 |
asb8
|
ankyrin repeat and SOCS box containing 8 |
| chr16_-_30826712 | 0.49 |
ENSDART00000122474
|
ptk2ab
|
protein tyrosine kinase 2ab |
| chr17_+_23753975 | 0.48 |
ENSDART00000135561
|
zgc:91976
|
zgc:91976 |
| chr18_+_36786842 | 0.48 |
ENSDART00000123264
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr16_-_42750295 | 0.48 |
ENSDART00000176570
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr7_-_40578733 | 0.47 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr24_-_2423791 | 0.47 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr6_+_23026170 | 0.47 |
ENSDART00000186683
|
srp68
|
signal recognition particle 68 |
| chr3_-_42016693 | 0.47 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr3_+_60607241 | 0.47 |
ENSDART00000167512
|
mfsd11
|
major facilitator superfamily domain containing 11 |
| chr20_-_54075136 | 0.47 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr22_-_18749313 | 0.47 |
ENSDART00000167466
|
midn
|
midnolin |
| chr11_-_25853212 | 0.46 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr16_-_7793457 | 0.46 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr14_+_7699443 | 0.46 |
ENSDART00000123139
|
brd8
|
bromodomain containing 8 |
| chr6_-_3978919 | 0.46 |
ENSDART00000167753
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr5_+_68807170 | 0.45 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr3_+_56574623 | 0.45 |
ENSDART00000130877
|
rac1b
|
Rac family small GTPase 1b |
| chr3_-_25086986 | 0.45 |
ENSDART00000050245
|
xpnpep3
|
X-prolyl aminopeptidase 3, mitochondrial |
| chr5_+_36655522 | 0.45 |
ENSDART00000015240
|
capns1a
|
calpain, small subunit 1 a |
| chr13_-_8446341 | 0.45 |
ENSDART00000080382
|
epas1b
|
endothelial PAS domain protein 1b |
| chr10_-_35149513 | 0.44 |
ENSDART00000063434
ENSDART00000131291 |
ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr2_+_5843694 | 0.44 |
ENSDART00000182364
|
dis3l2
|
DIS3 like 3'-5' exoribonuclease 2 |
| chr4_-_4834347 | 0.44 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx5a+tbx5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.4 | 1.1 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 0.9 | GO:0090008 | hypoblast development(GO:0090008) |
| 0.2 | 1.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 1.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.2 | 1.0 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.2 | 0.9 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.2 | 0.5 | GO:0001120 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.2 | 0.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.2 | 0.8 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.2 | 0.9 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.7 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.1 | 1.2 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 1.0 | GO:2000178 | negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.1 | 0.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.6 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.1 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.5 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.1 | 0.9 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 0.7 | GO:0032447 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.7 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 1.0 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 2.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.5 | GO:0055024 | regulation of cardiac muscle tissue development(GO:0055024) |
| 0.1 | 2.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.3 | GO:1902767 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.8 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.6 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.5 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.5 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.1 | 0.4 | GO:0048241 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.3 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.1 | 1.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 1.2 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 1.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.4 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 0.7 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.6 | GO:0002090 | regulation of receptor internalization(GO:0002090) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.5 | GO:0090497 | mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.8 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.2 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.1 | 1.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 4.3 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 0.6 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.1 | 0.6 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.4 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.1 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.1 | 0.9 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.7 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.2 | GO:2000389 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.4 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 1.1 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0070734 | histone H3-K27 methylation(GO:0070734) |
| 0.0 | 0.2 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.0 | 0.6 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.4 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.4 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0002857 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.4 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:0021611 | facial nerve formation(GO:0021611) |
| 0.0 | 0.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.5 | GO:0070307 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.5 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.7 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.4 | GO:0060005 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.8 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.7 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0045932 | negative regulation of muscle contraction(GO:0045932) |
| 0.0 | 0.1 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0097688 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.0 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.9 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.5 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.6 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.3 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.6 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 1.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.0 | 0.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.7 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.4 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.5 | GO:0009408 | response to heat(GO:0009408) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.6 | GO:0050870 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.4 | GO:0042761 | very long-chain fatty acid metabolic process(GO:0000038) very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.6 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.4 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.5 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.2 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.6 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.5 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.8 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.5 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.3 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.4 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 1.3 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 1.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 2.0 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.1 | GO:0055014 | atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.5 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.3 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0031055 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.4 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.1 | GO:0035678 | neuromast hair cell morphogenesis(GO:0035678) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.2 | 1.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.5 | GO:0030689 | Noc complex(GO:0030689) |
| 0.2 | 0.6 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 0.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.4 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.5 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 1.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.4 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 0.4 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 0.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.4 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.9 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 0.5 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.1 | 1.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.5 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.1 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.6 | GO:0030904 | retromer complex(GO:0030904) |
| 0.1 | 1.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.5 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 2.5 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.0 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.6 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.7 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.4 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.4 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 2.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 3.9 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0051139 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.2 | 1.2 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.8 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.2 | 1.7 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.9 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.4 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 1.3 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 1.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 1.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.5 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) |
| 0.1 | 2.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 2.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 2.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.5 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 0.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.8 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.1 | 1.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 0.2 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.1 | 0.8 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.5 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.4 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.6 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.9 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 0.6 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 2.8 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.8 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.9 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.1 | 0.5 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.5 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.7 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.5 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.4 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.6 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.5 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.5 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.6 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.7 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.8 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.5 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.1 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 1.1 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.1 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 1.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.0 | GO:1901612 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 1.2 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 1.0 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.9 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 1.8 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.6 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.5 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |