Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
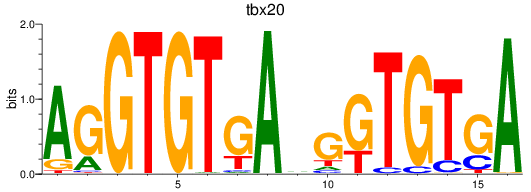
Results for tbx20
Z-value: 0.56
Transcription factors associated with tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx20
|
ENSDARG00000005150 | T-box transcription factor 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx20 | dr11_v1_chr16_-_42523744_42523744 | -0.60 | 7.9e-03 | Click! |
Activity profile of tbx20 motif
Sorted Z-values of tbx20 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_49537646 | 1.27 |
ENSDART00000180438
|
FO704848.1
|
|
| chr12_+_4920451 | 1.15 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr13_+_34690158 | 1.04 |
ENSDART00000182978
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr12_+_48340133 | 1.00 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr23_-_29668286 | 0.96 |
ENSDART00000129248
|
clstn1
|
calsyntenin 1 |
| chr11_-_26832685 | 0.89 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr21_-_5831413 | 0.89 |
ENSDART00000150914
|
wu:fj64h06
|
wu:fj64h06 |
| chr15_-_30867594 | 0.87 |
ENSDART00000154448
|
nf1a
|
neurofibromin 1a |
| chr2_-_57076687 | 0.85 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr7_+_40081630 | 0.84 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr9_-_32912638 | 0.80 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr8_-_42677086 | 0.77 |
ENSDART00000191173
|
si:ch73-138n13.1
|
si:ch73-138n13.1 |
| chr13_+_34689663 | 0.77 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr15_-_41689684 | 0.74 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr11_+_45233348 | 0.70 |
ENSDART00000173150
|
tmc6b
|
transmembrane channel-like 6b |
| chr21_-_12749501 | 0.68 |
ENSDART00000179724
|
LO018011.1
|
|
| chr14_-_33858214 | 0.67 |
ENSDART00000112268
|
ocrl
|
oculocerebrorenal syndrome of Lowe |
| chr15_-_47479119 | 0.67 |
ENSDART00000164957
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr25_+_18475032 | 0.66 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr20_+_42246948 | 0.64 |
ENSDART00000061135
|
gopc
|
golgi-associated PDZ and coiled-coil motif containing |
| chr10_+_42589707 | 0.61 |
ENSDART00000075269
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr22_-_37796998 | 0.58 |
ENSDART00000124742
ENSDART00000191232 |
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr17_-_11357851 | 0.55 |
ENSDART00000153915
|
si:ch211-185a18.2
|
si:ch211-185a18.2 |
| chr4_-_18840487 | 0.51 |
ENSDART00000066978
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr13_-_48431766 | 0.50 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr6_-_18751696 | 0.49 |
ENSDART00000171537
|
tnrc6c2
|
trinucleotide repeat containing 6C2 |
| chr7_-_40082053 | 0.49 |
ENSDART00000083719
ENSDART00000173687 |
shrprbck1r
|
sharpin and rbck1 related |
| chr21_+_1119046 | 0.44 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr4_-_18840919 | 0.39 |
ENSDART00000015834
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr12_+_1286642 | 0.39 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr20_+_32756763 | 0.38 |
ENSDART00000023006
|
fam84a
|
family with sequence similarity 84, member A |
| chr2_-_37797577 | 0.38 |
ENSDART00000110781
|
nfatc4
|
nuclear factor of activated T cells 4 |
| chr23_-_18381361 | 0.37 |
ENSDART00000016891
|
hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr17_+_1496107 | 0.36 |
ENSDART00000187804
|
LO018430.1
|
|
| chr11_+_45299447 | 0.29 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr23_-_27479558 | 0.29 |
ENSDART00000013563
|
atf7a
|
activating transcription factor 7a |
| chr24_-_41220538 | 0.26 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr23_+_21380079 | 0.26 |
ENSDART00000089379
|
iffo2a
|
intermediate filament family orphan 2a |
| chr12_-_43428542 | 0.23 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr9_+_23900703 | 0.23 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr13_-_10620652 | 0.21 |
ENSDART00000135000
ENSDART00000191587 |
si:ch73-54n14.2
camkmt
|
si:ch73-54n14.2 calmodulin-lysine N-methyltransferase |
| chr15_+_33696559 | 0.21 |
ENSDART00000188248
ENSDART00000161951 |
hdac12
|
histone deacetylase 12 |
| chr12_+_17933775 | 0.20 |
ENSDART00000186047
ENSDART00000160586 |
trrap
|
transformation/transcription domain-associated protein |
| chr5_+_30759232 | 0.18 |
ENSDART00000078018
|
ruvbl2
|
RuvB-like AAA ATPase 2 |
| chr12_+_15290800 | 0.17 |
ENSDART00000145656
|
med1
|
mediator complex subunit 1 |
| chr8_-_11112058 | 0.17 |
ENSDART00000042755
|
ampd1
|
adenosine monophosphate deaminase 1 (isoform M) |
| chr15_-_31516558 | 0.13 |
ENSDART00000156427
ENSDART00000156072 ENSDART00000156047 |
hmgb1b
|
high mobility group box 1b |
| chr25_-_7686201 | 0.11 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr3_-_47036951 | 0.11 |
ENSDART00000186971
|
CR392045.1
|
|
| chr24_+_42948 | 0.07 |
ENSDART00000122785
|
tmx3b
|
thioredoxin related transmembrane protein 3b |
| chr7_+_69449814 | 0.07 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr17_+_26965351 | 0.06 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr18_+_3572314 | 0.06 |
ENSDART00000169814
ENSDART00000157819 |
serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr25_+_36152215 | 0.05 |
ENSDART00000036147
|
irx5b
|
iroquois homeobox 5b |
| chr3_+_25857008 | 0.04 |
ENSDART00000154899
ENSDART00000193762 |
zgc:171844
|
zgc:171844 |
| chr3_-_8285123 | 0.04 |
ENSDART00000158699
ENSDART00000138588 |
trim35-9
|
tripartite motif containing 35-9 |
| chr4_-_50207986 | 0.04 |
ENSDART00000150388
|
si:ch211-197e7.3
|
si:ch211-197e7.3 |
| chr2_-_31301929 | 0.04 |
ENSDART00000191992
ENSDART00000190723 |
adcyap1b
|
adenylate cyclase activating polypeptide 1b |
| chr12_-_48127985 | 0.04 |
ENSDART00000193805
|
npffr1
|
neuropeptide FF receptor 1 |
| chr4_-_75812937 | 0.03 |
ENSDART00000125096
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr15_+_1765988 | 0.03 |
ENSDART00000159032
|
cul3b
|
cullin 3b |
| chr2_+_43469241 | 0.03 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr20_-_31808779 | 0.02 |
ENSDART00000133788
|
stxbp5a
|
syntaxin binding protein 5a (tomosyn) |
| chr17_+_33496067 | 0.02 |
ENSDART00000184724
|
pth2
|
parathyroid hormone 2 |
| chr22_-_35385810 | 0.02 |
ENSDART00000111824
|
HTR3C
|
5-hydroxytryptamine receptor 3C |
| chr8_+_36416285 | 0.01 |
ENSDART00000158400
|
si:busm1-194e12.8
|
si:busm1-194e12.8 |
| chr21_-_2273244 | 0.01 |
ENSDART00000171417
|
si:ch73-299h12.3
|
si:ch73-299h12.3 |
| chr7_+_39389273 | 0.01 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr25_+_19170500 | 0.01 |
ENSDART00000182572
|
BX294110.2
|
|
| chr4_+_35138928 | 0.01 |
ENSDART00000167934
ENSDART00000192116 ENSDART00000183689 |
si:dkey-279j5.1
|
si:dkey-279j5.1 |
| chr16_+_6750756 | 0.01 |
ENSDART00000149720
|
znf236
|
zinc finger protein 236 |
| chr13_-_293250 | 0.01 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr6_+_7555614 | 0.00 |
ENSDART00000187590
|
myh10
|
myosin, heavy chain 10, non-muscle |
| chr7_-_5070794 | 0.00 |
ENSDART00000097877
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr16_-_563732 | 0.00 |
ENSDART00000183394
|
irx2a
|
iroquois homeobox 2a |
| chr3_-_7940522 | 0.00 |
ENSDART00000159710
ENSDART00000167201 |
trim35-25
|
tripartite motif containing 35-25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx20
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 0.6 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.7 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.9 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.5 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 0.7 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.4 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.3 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.5 | GO:0060211 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.0 | 0.9 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0060420 | box C/D snoRNP assembly(GO:0000492) regulation of heart growth(GO:0060420) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.9 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.6 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.5 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.9 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.1 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.8 | GO:0051604 | protein maturation(GO:0051604) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 0.7 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.6 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0001163 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.3 | 0.9 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 1.0 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.7 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |