Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for tbx15_tbx1_mgaa_tbx4
Z-value: 0.84
Transcription factors associated with tbx15_tbx1_mgaa_tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
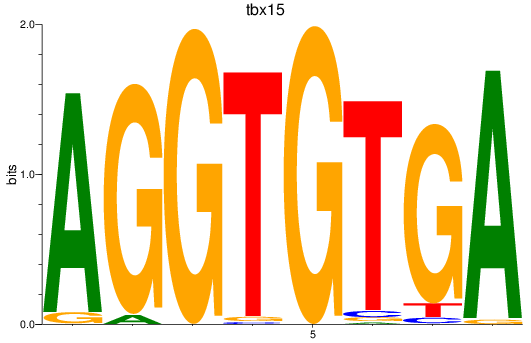
tbx15
|
ENSDARG00000002582 | T-box transcription factor 15 |
|
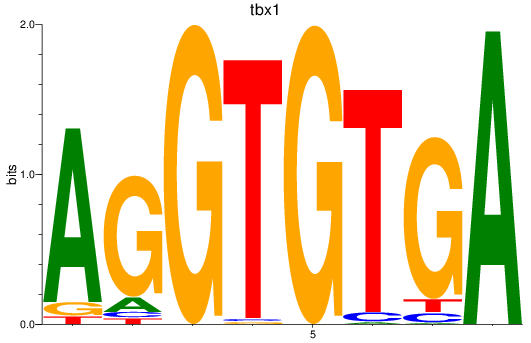
tbx1
|
ENSDARG00000031891 | T-box transcription factor 1 |
|
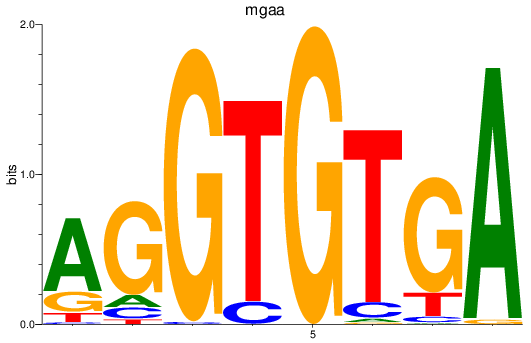
mgaa
|
ENSDARG00000078784 | MAX dimerization protein MGA a |
|
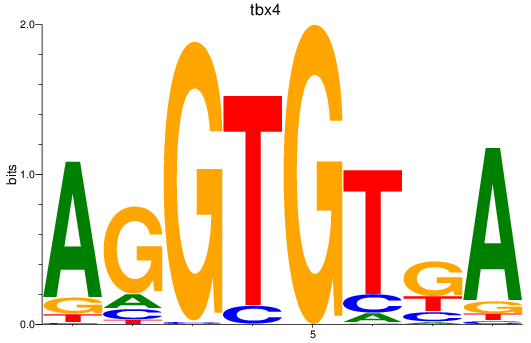
tbx4
|
ENSDARG00000030058 | T-box transcription factor 4 |
|
tbx4
|
ENSDARG00000113067 | T-box transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| mgaa | dr11_v1_chr17_+_10570906_10570906 | 0.89 | 7.2e-07 | Click! |
| tbx1 | dr11_v1_chr5_+_15203421_15203421 | 0.22 | 3.8e-01 | Click! |
| tbx4 | dr11_v1_chr15_+_27387555_27387555 | -0.20 | 4.2e-01 | Click! |
| tbx15 | dr11_v1_chr9_-_21067673_21067673 | 0.08 | 7.7e-01 | Click! |
Activity profile of tbx15_tbx1_mgaa_tbx4 motif
Sorted Z-values of tbx15_tbx1_mgaa_tbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_3700334 | 2.56 |
ENSDART00000137844
|
atp8b1
|
ATPase phospholipid transporting 8B1 |
| chr19_-_5332784 | 1.82 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr9_-_32912638 | 1.64 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr20_+_21268795 | 1.28 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr22_+_26600834 | 1.16 |
ENSDART00000157411
|
adcy9
|
adenylate cyclase 9 |
| chr1_-_52505279 | 1.11 |
ENSDART00000052907
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr2_+_36007449 | 1.06 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr22_-_3152357 | 1.05 |
ENSDART00000170983
|
lmnb2
|
lamin B2 |
| chr12_-_5448993 | 1.05 |
ENSDART00000181802
|
tbc1d12b
|
TBC1 domain family, member 12b |
| chr1_-_40341306 | 1.00 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr22_+_15959844 | 0.99 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr20_-_38778479 | 0.94 |
ENSDART00000185599
|
dnajc5ga
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a |
| chr23_-_10175898 | 0.93 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr22_+_15960005 | 0.92 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr1_-_53625142 | 0.91 |
ENSDART00000166852
|
USP34
|
ubiquitin specific peptidase 34 |
| chr12_-_35386910 | 0.91 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr11_-_3535537 | 0.89 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr16_+_16265110 | 0.86 |
ENSDART00000191492
|
setd2
|
SET domain containing 2 |
| chr7_-_28647959 | 0.86 |
ENSDART00000150148
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr14_+_14841685 | 0.83 |
ENSDART00000158291
ENSDART00000162039 |
slbp
|
stem-loop binding protein |
| chr21_+_15709061 | 0.80 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
| chr11_-_25257045 | 0.79 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr8_+_10862353 | 0.78 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr13_-_24880525 | 0.77 |
ENSDART00000136624
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr24_-_2423791 | 0.75 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr13_-_24906307 | 0.75 |
ENSDART00000148191
ENSDART00000189810 |
kat6b
|
K(lysine) acetyltransferase 6B |
| chr22_+_23430688 | 0.74 |
ENSDART00000160457
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr2_+_30531726 | 0.73 |
ENSDART00000146518
|
ankrd33bb
|
ankyrin repeat domain 33Bb |
| chr9_-_28255029 | 0.71 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr8_-_18225968 | 0.70 |
ENSDART00000135504
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr19_+_32456974 | 0.69 |
ENSDART00000088265
|
atxn1a
|
ataxin 1a |
| chr7_+_17947217 | 0.68 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr7_+_39418869 | 0.67 |
ENSDART00000169195
|
CT030188.1
|
|
| chr7_+_18100996 | 0.66 |
ENSDART00000055810
|
rab1ba
|
zRAB1B, member RAS oncogene family a |
| chr3_-_30888415 | 0.63 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr25_+_37397031 | 0.62 |
ENSDART00000193643
ENSDART00000169132 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr16_-_13613475 | 0.62 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr21_-_5831413 | 0.61 |
ENSDART00000150914
|
wu:fj64h06
|
wu:fj64h06 |
| chr15_-_21669618 | 0.61 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr8_-_1838315 | 0.61 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr16_+_50741154 | 0.61 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr9_+_34334156 | 0.61 |
ENSDART00000144272
|
pou2f1b
|
POU class 2 homeobox 1b |
| chr3_-_13955878 | 0.60 |
ENSDART00000166804
|
gcdhb
|
glutaryl-CoA dehydrogenase b |
| chr14_+_50937757 | 0.60 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr21_-_21178410 | 0.60 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr11_-_26832685 | 0.60 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr11_-_25257595 | 0.59 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr11_-_11910225 | 0.59 |
ENSDART00000159922
|
si:ch211-69b7.6
|
si:ch211-69b7.6 |
| chr17_+_26965351 | 0.59 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr7_-_39378903 | 0.56 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr16_-_35532937 | 0.56 |
ENSDART00000193209
|
ctps1b
|
CTP synthase 1b |
| chr22_-_4439311 | 0.56 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr12_-_17147473 | 0.55 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr19_+_791538 | 0.53 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr4_+_9028819 | 0.53 |
ENSDART00000102893
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr14_+_10954345 | 0.53 |
ENSDART00000106658
|
zdhhc15b
|
zinc finger, DHHC-type containing 15b |
| chr10_+_43994471 | 0.53 |
ENSDART00000138242
ENSDART00000186359 |
cldn5b
|
claudin 5b |
| chr6_-_16394528 | 0.52 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr5_+_60919378 | 0.52 |
ENSDART00000184915
|
doc2b
|
double C2-like domains, beta |
| chr12_+_32292564 | 0.51 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr6_-_49537646 | 0.51 |
ENSDART00000180438
|
FO704848.1
|
|
| chr7_+_31120766 | 0.51 |
ENSDART00000173703
|
tjp1a
|
tight junction protein 1a |
| chr13_-_8304605 | 0.51 |
ENSDART00000080460
|
atl2
|
atlastin GTPase 2 |
| chr13_+_21677767 | 0.50 |
ENSDART00000165166
|
mtg1
|
mitochondrial ribosome-associated GTPase 1 |
| chr22_+_15960514 | 0.50 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr7_+_39410180 | 0.49 |
ENSDART00000168641
|
CT030188.1
|
|
| chr11_+_24729346 | 0.48 |
ENSDART00000087740
|
zgc:153953
|
zgc:153953 |
| chr8_-_11229523 | 0.48 |
ENSDART00000002164
|
unc45b
|
unc-45 myosin chaperone B |
| chr3_-_15451097 | 0.46 |
ENSDART00000163836
|
BX784026.1
|
Danio rerio linker for activation of T cells (lat), mRNA. |
| chr17_+_58211 | 0.46 |
ENSDART00000157642
|
si:ch1073-209e23.1
|
si:ch1073-209e23.1 |
| chr5_+_44654535 | 0.46 |
ENSDART00000182190
ENSDART00000181872 |
dapk1
|
death-associated protein kinase 1 |
| chr7_+_39410393 | 0.46 |
ENSDART00000158561
ENSDART00000185173 |
CT030188.1
|
|
| chr13_+_25364324 | 0.45 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr8_+_49092077 | 0.45 |
ENSDART00000032355
|
zgc:56525
|
zgc:56525 |
| chr24_-_32025637 | 0.45 |
ENSDART00000180448
ENSDART00000159034 |
rsu1
|
Ras suppressor protein 1 |
| chr14_-_7245971 | 0.45 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr5_-_22615087 | 0.45 |
ENSDART00000146035
|
zgc:113208
|
zgc:113208 |
| chr18_+_14307059 | 0.44 |
ENSDART00000186558
|
zgc:173742
|
zgc:173742 |
| chr24_+_19518303 | 0.44 |
ENSDART00000027022
ENSDART00000056080 |
sulf1
|
sulfatase 1 |
| chr10_+_26944418 | 0.44 |
ENSDART00000135493
|
frmd8
|
FERM domain containing 8 |
| chr25_-_17918536 | 0.43 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr18_+_16053455 | 0.43 |
ENSDART00000189163
ENSDART00000188269 |
FO834850.1
|
|
| chr5_-_37959874 | 0.43 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr8_+_10869183 | 0.43 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr1_-_23110740 | 0.42 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr9_-_14992730 | 0.42 |
ENSDART00000137117
|
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr9_-_41323746 | 0.41 |
ENSDART00000140564
|
glsb
|
glutaminase b |
| chr13_-_27653679 | 0.41 |
ENSDART00000142568
|
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr24_+_19518570 | 0.41 |
ENSDART00000056081
|
sulf1
|
sulfatase 1 |
| chr1_-_31515746 | 0.41 |
ENSDART00000190886
|
cenpk
|
centromere protein K |
| chr17_-_26537928 | 0.41 |
ENSDART00000155692
ENSDART00000122366 |
ccser2a
|
coiled-coil serine-rich protein 2a |
| chr25_-_19666107 | 0.40 |
ENSDART00000149889
|
atp2b1b
|
ATPase plasma membrane Ca2+ transporting 1b |
| chr7_+_51795667 | 0.40 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr12_+_34770531 | 0.40 |
ENSDART00000153320
|
slc38a10
|
solute carrier family 38, member 10 |
| chr18_+_6536293 | 0.40 |
ENSDART00000024576
|
fkbp4
|
FK506 binding protein 4 |
| chr25_+_7321675 | 0.39 |
ENSDART00000104712
ENSDART00000142934 |
hmg20a
|
high mobility group 20A |
| chr10_-_31805923 | 0.39 |
ENSDART00000077785
|
vps26bl
|
vacuolar protein sorting 26 homolog B, like |
| chr2_-_6373829 | 0.39 |
ENSDART00000081633
|
si:dkey-119f1.1
|
si:dkey-119f1.1 |
| chr21_+_38073210 | 0.39 |
ENSDART00000186309
ENSDART00000142106 |
klf8
|
Kruppel-like factor 8 |
| chr25_+_418932 | 0.38 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr19_-_7420867 | 0.38 |
ENSDART00000081741
|
rab25a
|
RAB25, member RAS oncogene family a |
| chr14_+_28518349 | 0.38 |
ENSDART00000159961
|
stag2b
|
stromal antigen 2b |
| chr15_-_25269028 | 0.38 |
ENSDART00000078230
ENSDART00000193872 |
mettl16
|
methyltransferase like 16 |
| chr13_+_25364753 | 0.38 |
ENSDART00000027428
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr6_+_68220 | 0.38 |
ENSDART00000082955
|
sowahcb
|
sosondowah ankyrin repeat domain family Cb |
| chr1_-_34447515 | 0.38 |
ENSDART00000143048
|
lmo7b
|
LIM domain 7b |
| chr15_-_23482088 | 0.37 |
ENSDART00000185823
ENSDART00000185523 |
nlrx1
|
NLR family member X1 |
| chr19_-_31007417 | 0.37 |
ENSDART00000048144
|
rbbp4
|
retinoblastoma binding protein 4 |
| chr21_+_26726936 | 0.37 |
ENSDART00000065392
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr6_+_36877968 | 0.36 |
ENSDART00000155187
|
traf3ip2l
|
TRAF3 interacting protein 2-like |
| chr23_-_46020226 | 0.36 |
ENSDART00000160010
|
SYDE2
|
synapse defective Rho GTPase homolog 2 |
| chr15_-_18432673 | 0.35 |
ENSDART00000146853
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr17_+_51517750 | 0.35 |
ENSDART00000180896
ENSDART00000193528 |
pxdn
|
peroxidasin |
| chr5_+_44655148 | 0.35 |
ENSDART00000124059
|
dapk1
|
death-associated protein kinase 1 |
| chr18_+_6536598 | 0.34 |
ENSDART00000149350
|
fkbp4
|
FK506 binding protein 4 |
| chr17_-_10738001 | 0.34 |
ENSDART00000051526
|
jmjd7
|
jumonji domain containing 7 |
| chr13_-_36535128 | 0.34 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr9_-_21970067 | 0.34 |
ENSDART00000009920
|
lmo7a
|
LIM domain 7a |
| chr6_-_3924723 | 0.33 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr13_-_15928934 | 0.33 |
ENSDART00000142732
|
ttl
|
tubulin tyrosine ligase |
| chr16_+_20496691 | 0.33 |
ENSDART00000182737
ENSDART00000078984 |
cpvl
|
carboxypeptidase, vitellogenic-like |
| chr7_-_18656069 | 0.33 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr19_+_43669122 | 0.32 |
ENSDART00000139151
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr3_+_33367954 | 0.32 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr22_-_29640181 | 0.32 |
ENSDART00000059882
|
shoc2
|
SHOC2 leucine-rich repeat scaffold protein |
| chr1_+_45925150 | 0.32 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr7_+_29044888 | 0.31 |
ENSDART00000086871
|
gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr11_-_40647190 | 0.31 |
ENSDART00000173217
ENSDART00000173276 ENSDART00000147264 |
fam213b
|
family with sequence similarity 213, member B |
| chr6_-_3924543 | 0.31 |
ENSDART00000170584
|
tlk1b
|
tousled-like kinase 1b |
| chr21_+_10866421 | 0.31 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr25_-_13659249 | 0.31 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr2_+_33368414 | 0.31 |
ENSDART00000077462
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_-_10838434 | 0.30 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr23_+_27779452 | 0.30 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr20_+_41549200 | 0.29 |
ENSDART00000135715
|
fam184a
|
family with sequence similarity 184, member A |
| chr13_+_33368140 | 0.29 |
ENSDART00000033848
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr16_-_26676685 | 0.29 |
ENSDART00000103431
|
esrp1
|
epithelial splicing regulatory protein 1 |
| chr7_+_27455321 | 0.28 |
ENSDART00000148417
|
cyp2r1
|
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr1_+_37195465 | 0.28 |
ENSDART00000043855
ENSDART00000192580 ENSDART00000181666 |
dclk2a
|
doublecortin-like kinase 2a |
| chr21_-_26715270 | 0.28 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
| chr23_+_44644911 | 0.28 |
ENSDART00000140799
|
zgc:85858
|
zgc:85858 |
| chr17_+_31739418 | 0.28 |
ENSDART00000155073
ENSDART00000156180 |
arhgap5
|
Rho GTPase activating protein 5 |
| chr22_-_34872533 | 0.27 |
ENSDART00000167176
|
slit1b
|
slit homolog 1b (Drosophila) |
| chr12_-_10567188 | 0.27 |
ENSDART00000144283
|
myof
|
myoferlin |
| chr16_+_13822137 | 0.27 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr12_-_4475890 | 0.27 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr8_-_48847772 | 0.27 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr15_-_5580093 | 0.27 |
ENSDART00000143726
|
wdr62
|
WD repeat domain 62 |
| chr10_-_2788668 | 0.27 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr23_-_27442544 | 0.27 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr5_+_41146786 | 0.27 |
ENSDART00000175766
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr23_+_12840080 | 0.26 |
ENSDART00000081016
ENSDART00000121697 |
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr10_-_31782616 | 0.26 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr2_-_42552666 | 0.26 |
ENSDART00000141399
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr7_-_5431841 | 0.26 |
ENSDART00000173073
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr20_-_3238110 | 0.26 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr6_+_32326074 | 0.26 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr19_-_5812319 | 0.26 |
ENSDART00000114472
|
si:ch211-264f5.8
|
si:ch211-264f5.8 |
| chr10_+_8437930 | 0.26 |
ENSDART00000074553
|
pptc7b
|
PTC7 protein phosphatase homolog b |
| chr19_+_32979331 | 0.26 |
ENSDART00000078066
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr2_-_7185460 | 0.26 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr25_-_10610961 | 0.26 |
ENSDART00000153474
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr20_+_23676649 | 0.25 |
ENSDART00000035239
|
nek1
|
NIMA-related kinase 1 |
| chr19_+_13375838 | 0.25 |
ENSDART00000163093
|
lrp12
|
low density lipoprotein receptor-related protein 12 |
| chr3_-_42016693 | 0.25 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr19_-_42588510 | 0.25 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr12_-_26383242 | 0.25 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr9_+_33340311 | 0.25 |
ENSDART00000140064
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr24_-_36095526 | 0.25 |
ENSDART00000158145
|
CABZ01075509.1
|
|
| chr7_-_42206720 | 0.25 |
ENSDART00000110907
|
itfg1
|
integrin alpha FG-GAP repeat containing 1 |
| chr12_+_15290800 | 0.25 |
ENSDART00000145656
|
med1
|
mediator complex subunit 1 |
| chr5_+_19343880 | 0.25 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr22_-_14475927 | 0.25 |
ENSDART00000135768
|
lrp1ba
|
low density lipoprotein receptor-related protein 1Ba |
| chr1_-_49947290 | 0.25 |
ENSDART00000141476
|
sgms2
|
sphingomyelin synthase 2 |
| chr23_-_29667716 | 0.24 |
ENSDART00000158302
ENSDART00000133902 |
clstn1
|
calsyntenin 1 |
| chr10_-_44411032 | 0.24 |
ENSDART00000111509
|
CABZ01072096.1
|
|
| chr22_+_14117078 | 0.24 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr10_+_1052591 | 0.24 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr21_+_33503835 | 0.23 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr14_-_7207961 | 0.23 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr2_-_42396592 | 0.23 |
ENSDART00000127136
|
slco5a1b
|
solute carrier organic anion transporter family member 5A1b |
| chr11_+_18873113 | 0.23 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr12_+_48972915 | 0.23 |
ENSDART00000170695
|
lrit1b
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1b |
| chr18_-_13360106 | 0.23 |
ENSDART00000091512
|
cmip
|
c-Maf inducing protein |
| chr13_-_28610965 | 0.23 |
ENSDART00000043156
|
cyp17a1
|
cytochrome P450, family 17, subfamily A, polypeptide 1 |
| chr19_-_81851 | 0.23 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr2_-_42558549 | 0.23 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr2_+_15069011 | 0.23 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr22_-_16275236 | 0.23 |
ENSDART00000149051
|
cdc14ab
|
cell division cycle 14Ab |
| chr14_-_21660548 | 0.23 |
ENSDART00000161713
ENSDART00000089845 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr17_-_43031763 | 0.23 |
ENSDART00000132754
ENSDART00000050399 |
npc2
|
Niemann-Pick disease, type C2 |
| chr6_+_13232934 | 0.23 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr13_-_2283176 | 0.23 |
ENSDART00000158462
|
lrrc1
|
leucine rich repeat containing 1 |
| chr23_-_29667544 | 0.22 |
ENSDART00000059339
|
clstn1
|
calsyntenin 1 |
| chr20_-_26846028 | 0.22 |
ENSDART00000136687
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr8_+_47571211 | 0.22 |
ENSDART00000131460
|
plch2a
|
phospholipase C, eta 2a |
| chr15_+_42397125 | 0.22 |
ENSDART00000169751
|
tiam1b
|
T cell lymphoma invasion and metastasis 1b |
| chr4_-_60049792 | 0.22 |
ENSDART00000158199
|
znf1033
|
zinc finger protein 1033 |
| chr5_+_31811662 | 0.22 |
ENSDART00000023463
|
uap1l1
|
UDP-N-acetylglucosamine pyrophosphorylase 1, like 1 |
| chr18_-_11595567 | 0.22 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx15_tbx1_mgaa_tbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.2 | 0.7 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 | 0.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 1.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.2 | 0.5 | GO:0009258 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 0.8 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 0.9 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 0.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.6 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.1 | 0.8 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.5 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.9 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.3 | GO:0060092 | regulation of synaptic transmission, glycinergic(GO:0060092) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.6 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0030237 | female sex determination(GO:0030237) male sex determination(GO:0030238) |
| 0.1 | 2.6 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.4 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.1 | 0.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 1.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.9 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.2 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.2 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.2 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.1 | 0.2 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.1 | 0.3 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.5 | GO:0071295 | cellular response to nutrient(GO:0031670) cellular response to vitamin(GO:0071295) |
| 0.1 | 0.3 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.1 | 0.2 | GO:0001783 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 0.5 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.7 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.0 | 0.5 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.5 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.4 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) pancreas morphogenesis(GO:0061113) |
| 0.0 | 1.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.3 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.0 | 0.8 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.3 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.2 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0048662 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.9 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0006266 | DNA ligation(GO:0006266) immunoglobulin V(D)J recombination(GO:0033152) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.2 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.3 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.2 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.0 | 0.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.3 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.6 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.2 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.8 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.7 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.1 | GO:0000305 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.4 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 1.6 | GO:0031497 | chromatin assembly(GO:0031497) |
| 0.0 | 0.6 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.4 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.0 | 0.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.1 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.0 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.3 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.0 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 1.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.2 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.9 | GO:0009190 | cyclic nucleotide biosynthetic process(GO:0009190) |
| 0.0 | 0.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.5 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.3 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.1 | 0.8 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.4 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.1 | 1.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.4 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 2.8 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 3.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 2.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 2.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 2.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.4 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 2.2 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.2 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 0.8 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.2 | 0.6 | GO:0001164 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.2 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 0.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.1 | 0.6 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.6 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.4 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 1.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.6 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 1.2 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.1 | 0.3 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.9 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.5 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.8 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.0 | 0.3 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 1.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.2 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.9 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.2 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.6 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 1.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0070404 | 6,7-dihydropteridine reductase activity(GO:0004155) NADH binding(GO:0070404) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.0 | 0.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.6 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 1.4 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.6 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.7 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.8 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 2.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 0.8 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 0.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 0.4 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 2.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |