Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
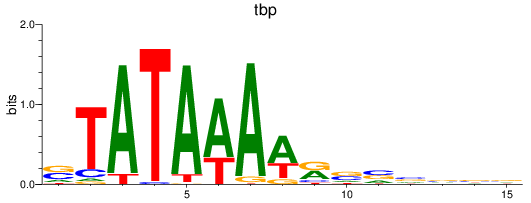
Results for tbp
Z-value: 1.29
Transcription factors associated with tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbp
|
ENSDARG00000014994 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbp | dr11_v1_chr13_-_24396199_24396199 | -0.78 | 1.2e-04 | Click! |
Activity profile of tbp motif
Sorted Z-values of tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_25085327 | 4.62 |
ENSDART00000077661
|
prss1
|
protease, serine 1 |
| chr6_+_269204 | 3.91 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr10_-_44924289 | 3.68 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr13_-_15702672 | 3.45 |
ENSDART00000144445
ENSDART00000168950 |
ckba
|
creatine kinase, brain a |
| chr2_-_38000276 | 3.11 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr5_+_37978501 | 3.03 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr9_-_1703761 | 2.85 |
ENSDART00000144822
ENSDART00000137210 ENSDART00000135273 |
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr5_-_71722257 | 2.47 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr2_+_55365727 | 2.39 |
ENSDART00000162943
|
FP245456.1
|
|
| chr5_-_27867657 | 2.28 |
ENSDART00000112495
|
tcima
|
transcriptional and immune response regulator a |
| chr18_+_25752592 | 2.27 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr11_+_6456146 | 2.26 |
ENSDART00000036939
|
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr7_+_35068036 | 2.17 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr21_+_7823146 | 2.08 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr5_+_4366431 | 2.00 |
ENSDART00000168560
ENSDART00000149185 |
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr24_-_41312459 | 1.99 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr8_-_31369161 | 1.92 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr15_+_28368823 | 1.89 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr6_+_49052741 | 1.83 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr4_+_842010 | 1.82 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr25_+_30298377 | 1.78 |
ENSDART00000153622
|
C11orf96
|
chromosome 11 open reading frame 96 |
| chr2_+_16780643 | 1.77 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr21_-_280769 | 1.75 |
ENSDART00000157753
|
plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr23_+_27675581 | 1.74 |
ENSDART00000127198
|
rps26
|
ribosomal protein S26 |
| chr15_-_5815006 | 1.73 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr3_+_15505275 | 1.73 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr22_+_16497670 | 1.71 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr5_-_22027357 | 1.69 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr3_-_24456451 | 1.68 |
ENSDART00000024480
ENSDART00000156814 |
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr23_-_35082494 | 1.68 |
ENSDART00000189809
|
BX294434.1
|
|
| chr16_-_23471387 | 1.62 |
ENSDART00000180899
|
CR533578.1
|
|
| chr25_+_22296666 | 1.61 |
ENSDART00000138887
ENSDART00000193410 |
cyp11a2
|
cytochrome P450, family 11, subfamily A, polypeptide 2 |
| chr18_-_50799510 | 1.60 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr22_+_18530395 | 1.58 |
ENSDART00000105415
ENSDART00000183958 |
si:ch211-212d10.1
|
si:ch211-212d10.1 |
| chr14_-_8724290 | 1.56 |
ENSDART00000161171
|
pimr56
|
Pim proto-oncogene, serine/threonine kinase, related 56 |
| chr6_-_54180699 | 1.54 |
ENSDART00000045901
|
rps10
|
ribosomal protein S10 |
| chr16_-_45917322 | 1.53 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr24_-_27419198 | 1.53 |
ENSDART00000141124
|
ccl34b.4
|
chemokine (C-C motif) ligand 34b, duplicate 4 |
| chr9_-_52386733 | 1.48 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr24_+_32176155 | 1.42 |
ENSDART00000003745
|
vim
|
vimentin |
| chr10_-_44027391 | 1.39 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr5_-_33259079 | 1.39 |
ENSDART00000132223
|
ifitm1
|
interferon induced transmembrane protein 1 |
| chr13_-_12660318 | 1.36 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr22_+_7480465 | 1.35 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr10_-_24371312 | 1.35 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr14_-_10617923 | 1.35 |
ENSDART00000133723
ENSDART00000131939 ENSDART00000136649 |
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr19_-_41069573 | 1.34 |
ENSDART00000111982
ENSDART00000193142 |
sgce
|
sarcoglycan, epsilon |
| chr23_+_19813677 | 1.34 |
ENSDART00000139192
ENSDART00000142308 |
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr14_-_28001986 | 1.32 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr20_-_53981626 | 1.32 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr3_-_57425961 | 1.30 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr15_-_34558579 | 1.30 |
ENSDART00000140882
|
ispd
|
isoprenoid synthase domain containing |
| chr25_-_32449235 | 1.29 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr7_+_34687969 | 1.27 |
ENSDART00000182013
ENSDART00000188255 ENSDART00000182728 |
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr25_-_12982193 | 1.24 |
ENSDART00000159617
|
ccl39.5
|
chemokine (C-C motif) ligand 39, duplicate 5 |
| chr4_+_306036 | 1.24 |
ENSDART00000103659
|
msgn1
|
mesogenin 1 |
| chr12_+_7254112 | 1.21 |
ENSDART00000092009
|
tfam
|
transcription factor A, mitochondrial |
| chr7_-_7823662 | 1.21 |
ENSDART00000167652
|
cxcl8b.3
|
chemokine (C-X-C motif) ligand 8b, duplicate 3 |
| chr21_+_25221940 | 1.20 |
ENSDART00000108972
|
sycn.1
|
syncollin, tandem duplicate 1 |
| chr2_+_2181709 | 1.20 |
ENSDART00000092763
ENSDART00000148553 |
ccdc13
|
coiled-coil domain containing 13 |
| chr11_+_13207898 | 1.15 |
ENSDART00000060310
|
atp5f1b
|
ATP synthase F1 subunit beta |
| chr24_+_21621654 | 1.14 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr10_-_22845485 | 1.14 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr23_+_25305431 | 1.13 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr22_+_18188045 | 1.09 |
ENSDART00000140106
|
mef2b
|
myocyte enhancer factor 2b |
| chr13_+_17694845 | 1.09 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr22_-_21392748 | 1.09 |
ENSDART00000144648
|
ankrd24
|
ankyrin repeat domain 24 |
| chr7_-_73851280 | 1.07 |
ENSDART00000190053
|
FP236812.3
|
|
| chr23_+_32406009 | 1.06 |
ENSDART00000155793
|
si:ch211-66i15.5
|
si:ch211-66i15.5 |
| chr14_-_33334065 | 1.05 |
ENSDART00000052761
|
rpl39
|
ribosomal protein L39 |
| chr8_+_26818446 | 1.05 |
ENSDART00000134987
ENSDART00000138835 |
si:ch211-156j16.1
|
si:ch211-156j16.1 |
| chr5_-_36837846 | 1.04 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr3_-_55121125 | 1.04 |
ENSDART00000125092
|
hbae1
|
hemoglobin, alpha embryonic 1 |
| chr22_+_5752257 | 1.02 |
ENSDART00000143052
|
cox14
|
COX14 cytochrome c oxidase assembly factor |
| chr6_-_346084 | 1.01 |
ENSDART00000162599
|
pde6ha
|
phosphodiesterase 6H, cGMP-specific, cone, gamma, paralog a |
| chr24_-_37688398 | 1.00 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr10_+_15777258 | 1.00 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr2_+_24536762 | 0.98 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr10_+_42374957 | 0.98 |
ENSDART00000147926
|
COX5B
|
zgc:86599 |
| chr2_+_16781015 | 0.96 |
ENSDART00000155147
ENSDART00000003845 |
tfa
|
transferrin-a |
| chr22_+_18187857 | 0.96 |
ENSDART00000166300
|
mef2b
|
myocyte enhancer factor 2b |
| chr2_+_35615547 | 0.94 |
ENSDART00000135843
|
ankrd45
|
ankyrin repeat domain 45 |
| chr1_-_49225890 | 0.94 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr13_+_35745572 | 0.92 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr25_-_18454016 | 0.92 |
ENSDART00000005877
|
cpa1
|
carboxypeptidase A1 (pancreatic) |
| chr2_-_30182353 | 0.92 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr8_-_37461835 | 0.89 |
ENSDART00000013446
|
eif2d
|
eukaryotic translation initiation factor 2D |
| chr19_-_32518556 | 0.88 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr3_+_26135502 | 0.87 |
ENSDART00000146979
|
atp2a1
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 1 |
| chr6_+_46431848 | 0.87 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr12_+_48204891 | 0.85 |
ENSDART00000190534
ENSDART00000164427 |
ndr2
|
nodal-related 2 |
| chr11_+_25472758 | 0.85 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr1_+_10683843 | 0.83 |
ENSDART00000054879
|
zgc:103678
|
zgc:103678 |
| chr6_+_3934738 | 0.82 |
ENSDART00000159673
|
dync1i2b
|
dynein, cytoplasmic 1, intermediate chain 2b |
| chr23_+_26979174 | 0.82 |
ENSDART00000018654
|
rnd1b
|
Rho family GTPase 1b |
| chr25_+_4979446 | 0.82 |
ENSDART00000154131
ENSDART00000155537 |
si:ch73-265h17.1
|
si:ch73-265h17.1 |
| chr19_+_24882845 | 0.81 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr21_+_25226558 | 0.81 |
ENSDART00000168480
|
sycn.2
|
syncollin, tandem duplicate 2 |
| chr5_-_67911111 | 0.80 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr12_-_19346678 | 0.79 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr3_-_36606884 | 0.79 |
ENSDART00000172003
|
si:dkeyp-72e1.6
|
si:dkeyp-72e1.6 |
| chr15_-_4967302 | 0.79 |
ENSDART00000101992
|
lipt2
|
lipoyl(octanoyl) transferase 2 |
| chr20_+_51813432 | 0.79 |
ENSDART00000127444
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr6_-_53426773 | 0.79 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr2_+_24507770 | 0.79 |
ENSDART00000154802
ENSDART00000052063 |
rps28
|
ribosomal protein S28 |
| chr3_-_32818607 | 0.77 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr11_-_39202915 | 0.77 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr5_+_68807170 | 0.77 |
ENSDART00000017849
|
her7
|
hairy and enhancer of split related-7 |
| chr10_+_41986685 | 0.77 |
ENSDART00000086165
ENSDART00000171983 |
setd1ba
|
SET domain containing 1B, a |
| chr21_-_12030654 | 0.77 |
ENSDART00000139145
|
tpgs2
|
tubulin polyglutamylase complex subunit 2 |
| chr18_+_30507839 | 0.76 |
ENSDART00000026866
|
cox4i1
|
cytochrome c oxidase subunit IV isoform 1 |
| chr25_+_8356707 | 0.76 |
ENSDART00000153708
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr3_-_23643751 | 0.76 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr13_-_2215213 | 0.76 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr5_+_37837245 | 0.75 |
ENSDART00000171617
|
epd
|
ependymin |
| chr2_-_127945 | 0.73 |
ENSDART00000056453
|
igfbp1b
|
insulin-like growth factor binding protein 1b |
| chr17_+_25397070 | 0.71 |
ENSDART00000164254
|
zgc:154055
|
zgc:154055 |
| chr5_-_26181863 | 0.71 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr5_-_41645058 | 0.70 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr4_-_8035520 | 0.70 |
ENSDART00000146146
|
si:ch211-240l19.7
|
si:ch211-240l19.7 |
| chr2_-_37837056 | 0.69 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr1_-_53880639 | 0.69 |
ENSDART00000010543
|
ltv1
|
LTV1 ribosome biogenesis factor |
| chr7_+_20524064 | 0.69 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr2_+_24507597 | 0.68 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr8_+_6863462 | 0.68 |
ENSDART00000064163
|
nefmb
|
neurofilament, medium polypeptide b |
| chr25_+_36329623 | 0.66 |
ENSDART00000073405
|
zgc:173552
|
zgc:173552 |
| chr8_+_999421 | 0.65 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr5_+_69733096 | 0.64 |
ENSDART00000169013
|
arl6ip4
|
ADP-ribosylation factor-like 6 interacting protein 4 |
| chr2_+_17181777 | 0.64 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr7_+_29951997 | 0.64 |
ENSDART00000173453
|
tpma
|
alpha-tropomyosin |
| chr22_-_11626014 | 0.64 |
ENSDART00000063133
ENSDART00000160085 |
gcga
|
glucagon a |
| chr6_-_53281518 | 0.63 |
ENSDART00000157621
|
rbm5
|
RNA binding motif protein 5 |
| chr5_-_32274383 | 0.63 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr15_+_714203 | 0.63 |
ENSDART00000153847
|
si:dkey-7i4.24
|
si:dkey-7i4.24 |
| chr12_+_48390715 | 0.62 |
ENSDART00000149351
|
scd
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr13_+_22295905 | 0.61 |
ENSDART00000180133
ENSDART00000181125 |
usp54a
|
ubiquitin specific peptidase 54a |
| chr4_-_20485508 | 0.61 |
ENSDART00000183276
ENSDART00000023337 |
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr7_-_73846995 | 0.61 |
ENSDART00000188079
|
FP236812.4
|
|
| chr6_+_39493864 | 0.60 |
ENSDART00000086263
|
mettl7a
|
methyltransferase like 7A |
| chr12_+_28799988 | 0.60 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr8_+_29856013 | 0.60 |
ENSDART00000061981
ENSDART00000149610 |
hsd17b3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr15_+_1134870 | 0.59 |
ENSDART00000155392
|
p2ry13
|
purinergic receptor P2Y13 |
| chr12_-_17712393 | 0.59 |
ENSDART00000143534
ENSDART00000010144 |
pvalb2
|
parvalbumin 2 |
| chr21_-_28523548 | 0.59 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr22_-_9934854 | 0.59 |
ENSDART00000136404
|
si:dkey-253d23.11
|
si:dkey-253d23.11 |
| chr21_+_30043054 | 0.59 |
ENSDART00000065448
|
fabp6
|
fatty acid binding protein 6, ileal (gastrotropin) |
| chr20_-_36227500 | 0.59 |
ENSDART00000166054
|
BX255920.1
|
|
| chr13_-_50624743 | 0.59 |
ENSDART00000167949
|
vox
|
ventral homeobox |
| chr24_+_37598441 | 0.58 |
ENSDART00000125145
|
rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila) |
| chr2_+_30904069 | 0.57 |
ENSDART00000017144
|
bag1
|
BCL2 associated athanogene 1 |
| chr4_-_74367912 | 0.57 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr5_-_10239079 | 0.56 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr17_+_890988 | 0.56 |
ENSDART00000186843
|
CABZ01078858.1
|
|
| chr16_-_45917683 | 0.56 |
ENSDART00000184289
|
afp4
|
antifreeze protein type IV |
| chr13_+_35746440 | 0.55 |
ENSDART00000187859
|
gpr75
|
G protein-coupled receptor 75 |
| chr3_+_5575313 | 0.55 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr6_+_35362225 | 0.55 |
ENSDART00000133783
ENSDART00000102483 |
rgs4
|
regulator of G protein signaling 4 |
| chr17_-_50234004 | 0.54 |
ENSDART00000058706
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr16_-_24672919 | 0.52 |
ENSDART00000008326
|
pon2
|
paraoxonase 2 |
| chr7_-_73845736 | 0.52 |
ENSDART00000193414
|
zgc:173552
|
zgc:173552 |
| chr20_-_39273987 | 0.52 |
ENSDART00000127173
|
clu
|
clusterin |
| chr8_+_30671060 | 0.51 |
ENSDART00000193749
|
adora2aa
|
adenosine A2a receptor a |
| chr10_-_5058823 | 0.51 |
ENSDART00000139825
|
tmem150c
|
transmembrane protein 150C |
| chr20_+_16721933 | 0.50 |
ENSDART00000063950
|
psmc1b
|
proteasome 26S subunit, ATPase 1b |
| chr6_+_39499623 | 0.49 |
ENSDART00000036057
|
si:ch211-173n18.3
|
si:ch211-173n18.3 |
| chr3_-_6709938 | 0.49 |
ENSDART00000172196
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr18_-_2668698 | 0.49 |
ENSDART00000157510
|
relt
|
RELT, TNF receptor |
| chr18_-_20532339 | 0.48 |
ENSDART00000060291
|
ighmbp2
|
immunoglobulin mu binding protein 2 |
| chr12_+_17042754 | 0.46 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr14_-_38828057 | 0.46 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr9_+_31075896 | 0.46 |
ENSDART00000188042
|
clybl
|
citrate lyase beta like |
| chr1_-_22338521 | 0.45 |
ENSDART00000176849
|
si:ch73-380n15.2
|
si:ch73-380n15.2 |
| chr8_-_6877390 | 0.45 |
ENSDART00000170883
ENSDART00000005321 |
neflb
|
neurofilament, light polypeptide b |
| chr4_+_25706037 | 0.45 |
ENSDART00000141133
|
lamb1b
|
laminin, beta 1b |
| chr13_-_20381485 | 0.45 |
ENSDART00000131351
|
si:ch211-270n8.1
|
si:ch211-270n8.1 |
| chr20_+_42668875 | 0.45 |
ENSDART00000048890
|
slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chr3_-_31804481 | 0.45 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr15_+_15390882 | 0.44 |
ENSDART00000062024
|
ca4b
|
carbonic anhydrase IV b |
| chr6_+_11250033 | 0.44 |
ENSDART00000065411
ENSDART00000132677 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr24_-_1303935 | 0.44 |
ENSDART00000159212
ENSDART00000159267 ENSDART00000164904 |
nrp1a
|
neuropilin 1a |
| chr8_+_23703464 | 0.43 |
ENSDART00000132584
|
ppardb
|
peroxisome proliferator-activated receptor delta b |
| chr5_-_7834582 | 0.43 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr23_+_44236281 | 0.42 |
ENSDART00000149842
|
MEPCE
|
si:ch1073-157b13.1 |
| chr11_+_34235372 | 0.42 |
ENSDART00000063150
|
fam43a
|
family with sequence similarity 43, member A |
| chr12_+_20641102 | 0.42 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr12_+_26632448 | 0.41 |
ENSDART00000185762
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr12_-_20665164 | 0.41 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr7_+_29153018 | 0.40 |
ENSDART00000138065
|
tmppe
|
transmembrane protein with metallophosphoesterase domain |
| chr6_+_11249706 | 0.40 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr9_+_23770666 | 0.40 |
ENSDART00000182493
|
si:ch211-219a4.3
|
si:ch211-219a4.3 |
| chr14_-_33872092 | 0.40 |
ENSDART00000111903
|
si:ch73-335m24.2
|
si:ch73-335m24.2 |
| chr10_-_7785930 | 0.39 |
ENSDART00000043961
ENSDART00000111058 |
mpx
|
myeloid-specific peroxidase |
| chr4_+_34343524 | 0.39 |
ENSDART00000171694
|
si:ch211-246b8.5
|
si:ch211-246b8.5 |
| chr15_+_509126 | 0.39 |
ENSDART00000102274
|
ftr86
|
finTRIM family, member 86 |
| chr21_-_23308286 | 0.39 |
ENSDART00000184419
|
zbtb16a
|
zinc finger and BTB domain containing 16a |
| chr15_+_45643787 | 0.39 |
ENSDART00000055995
ENSDART00000157750 |
sagb
|
S-antigen; retina and pineal gland (arrestin) b |
| chr4_-_4780667 | 0.39 |
ENSDART00000133973
|
si:ch211-258f14.2
|
si:ch211-258f14.2 |
| chr10_+_4964463 | 0.39 |
ENSDART00000109882
|
lyrm7
|
LYR motif containing 7 |
| chr15_-_23814330 | 0.38 |
ENSDART00000153843
|
si:ch211-167j9.5
|
si:ch211-167j9.5 |
| chr6_-_426041 | 0.38 |
ENSDART00000162789
|
fam83fb
|
family with sequence similarity 83, member Fb |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.6 | 1.8 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) chiasma assembly(GO:0051026) |
| 0.6 | 3.0 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.5 | 2.0 | GO:0006598 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.5 | 4.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 1.8 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.4 | 1.2 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.4 | 3.9 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.4 | 2.5 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.3 | 4.5 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.3 | 0.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.3 | 1.6 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.3 | 1.6 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.3 | 1.2 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.3 | 1.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.3 | 0.8 | GO:0072592 | oxygen metabolic process(GO:0072592) regulation of oxygen metabolic process(GO:2000374) positive regulation of oxygen metabolic process(GO:2000376) |
| 0.3 | 1.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.2 | 2.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.2 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 0.7 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 0.9 | GO:0031446 | regulation of twitch skeletal muscle contraction(GO:0014724) regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) negative regulation of striated muscle contraction(GO:0045988) relaxation of skeletal muscle(GO:0090076) |
| 0.2 | 1.3 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.2 | 1.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.6 | GO:0042364 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 1.3 | GO:0070571 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.2 | 1.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.6 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.2 | 0.6 | GO:0070291 | N-acylethanolamine metabolic process(GO:0070291) |
| 0.1 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.1 | 1.5 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 3.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.1 | GO:0048327 | axial mesodermal cell differentiation(GO:0048321) axial mesodermal cell fate commitment(GO:0048322) axial mesodermal cell fate specification(GO:0048327) |
| 0.1 | 0.5 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.6 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 1.0 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 1.0 | GO:0046619 | optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.1 | 0.4 | GO:0046888 | regulation of endocrine process(GO:0044060) negative regulation of hormone secretion(GO:0046888) endocrine hormone secretion(GO:0060986) |
| 0.1 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.7 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.3 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.1 | 5.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 1.0 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.1 | 0.4 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 2.2 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 1.1 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.2 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.2 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.1 | 0.3 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.1 | 0.3 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 2.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 0.7 | GO:0042304 | regulation of fatty acid biosynthetic process(GO:0042304) |
| 0.1 | 0.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 | 0.2 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.1 | 2.1 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.1 | 0.4 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.1 | 0.6 | GO:0046661 | male sex differentiation(GO:0046661) |
| 0.1 | 0.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.7 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.9 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 1.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.2 | GO:1902267 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 2.2 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.0 | 0.1 | GO:0016117 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.4 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.0 | 3.9 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.8 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.7 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.4 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.8 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 1.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.5 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.2 | GO:0032732 | interleukin-1 production(GO:0032612) regulation of interleukin-1 production(GO:0032652) positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.6 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.0 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 1.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 1.1 | GO:1901214 | regulation of neuron death(GO:1901214) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.4 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.5 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 0.5 | 1.8 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.3 | 3.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.3 | 0.8 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 3.7 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 1.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.2 | 1.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 0.8 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 2.1 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 4.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 0.8 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 3.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.9 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0032426 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) stereocilium tip(GO:0032426) |
| 0.0 | 0.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 17.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 1.3 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.6 | 3.9 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.5 | 1.6 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.4 | 1.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 4.5 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.3 | 1.2 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.3 | 1.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.3 | 2.0 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.3 | 1.6 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.3 | 0.8 | GO:0033819 | lipoyl(octanoyl) transferase activity(GO:0033819) |
| 0.2 | 0.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.1 | GO:0051903 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.2 | 1.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.2 | 1.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 2.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.2 | 0.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.2 | 0.6 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 0.5 | GO:0047777 | (3S)-citramalyl-CoA lyase activity(GO:0047777) |
| 0.2 | 0.5 | GO:0042166 | neurotransmitter binding(GO:0042165) acetylcholine binding(GO:0042166) |
| 0.1 | 0.5 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 0.9 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.8 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 1.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 1.6 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.1 | 0.5 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 2.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.1 | 0.5 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.7 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 1.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.1 | 3.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.1 | 0.6 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 2.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 0.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 4.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 1.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 9.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 7.9 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 1.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.0 | 0.9 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 1.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.1 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.5 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.6 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.6 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.7 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.6 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 3.9 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.1 | GO:0042936 | dipeptide transporter activity(GO:0042936) dipeptide transmembrane transporter activity(GO:0071916) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.8 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.9 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.4 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.6 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.4 | 1.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.2 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.2 | 1.4 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 4.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.6 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 4.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.9 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.3 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.2 | REACTOME RNA POL I RNA POL III AND MITOCHONDRIAL TRANSCRIPTION | Genes involved in RNA Polymerase I, RNA Polymerase III, and Mitochondrial Transcription |
| 0.0 | 3.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.6 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |