Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
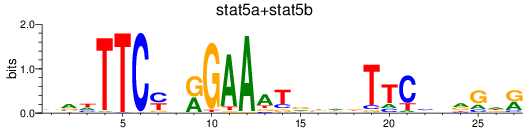
Results for stat5a+stat5b
Z-value: 0.62
Transcription factors associated with stat5a+stat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat5a
|
ENSDARG00000019392 | signal transducer and activator of transcription 5a |
|
stat5b
|
ENSDARG00000055588 | signal transducer and activator of transcription 5b |
|
stat5a
|
ENSDARG00000113871 | signal transducer and activator of transcription 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat5a | dr11_v1_chr3_+_17251542_17251542 | -0.76 | 2.9e-04 | Click! |
| stat5b | dr11_v1_chr12_-_13650344_13650472 | -0.56 | 1.7e-02 | Click! |
Activity profile of stat5a+stat5b motif
Sorted Z-values of stat5a+stat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_36914636 | 2.99 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr16_+_34523515 | 2.79 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr8_-_47212686 | 2.27 |
ENSDART00000040370
|
pimr189
|
Pim proto-oncogene, serine/threonine kinase, related 189 |
| chr3_+_55100045 | 2.14 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr8_-_46060685 | 2.13 |
ENSDART00000121902
|
pimr188
|
Pim proto-oncogene, serine/threonine kinase, related 188 |
| chr15_-_25083200 | 1.91 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr15_-_25010400 | 1.89 |
ENSDART00000155236
|
si:ch211-198e20.10
|
si:ch211-198e20.10 |
| chr11_-_17964525 | 1.65 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr15_-_25039083 | 1.64 |
ENSDART00000154409
|
pimr190
|
Pim proto-oncogene, serine/threonine kinase, related 190 |
| chr17_+_46864116 | 1.43 |
ENSDART00000156250
|
pimr27
|
Pim proto-oncogene, serine/threonine kinase, related 27 |
| chr22_+_7480465 | 1.40 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr16_+_25126935 | 1.31 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr18_-_48558420 | 1.23 |
ENSDART00000058987
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr17_-_24521382 | 1.14 |
ENSDART00000092948
|
peli1b
|
pellino E3 ubiquitin protein ligase 1b |
| chr11_+_30321116 | 1.14 |
ENSDART00000187921
ENSDART00000127075 |
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr6_+_16895685 | 1.06 |
ENSDART00000154090
|
pimr28
|
Pim proto-oncogene, serine/threonine kinase, related 28 |
| chr20_+_25586099 | 1.06 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr6_+_16870004 | 1.02 |
ENSDART00000154794
|
pimr21
|
Pim proto-oncogene, serine/threonine kinase, related 21 |
| chr6_+_45934682 | 0.97 |
ENSDART00000103489
|
cenps
|
centromere protein S |
| chr2_+_22659787 | 0.95 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr12_+_22680115 | 0.91 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr22_+_2937485 | 0.90 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr4_-_57572054 | 0.77 |
ENSDART00000158610
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr4_-_18595525 | 0.76 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr1_-_44025066 | 0.74 |
ENSDART00000148932
|
si:ch73-109d9.1
|
si:ch73-109d9.1 |
| chr17_-_46817295 | 0.69 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr16_+_9762261 | 0.69 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr17_+_12255642 | 0.68 |
ENSDART00000155175
|
pimr175
|
Pim proto-oncogene, serine/threonine kinase, related 175 |
| chr4_-_12323228 | 0.68 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr13_+_6092135 | 0.67 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr17_-_24439672 | 0.66 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr4_-_1497384 | 0.64 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr17_-_46933567 | 0.61 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr17_-_26604946 | 0.61 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr15_-_41290415 | 0.61 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr4_+_47174366 | 0.59 |
ENSDART00000170732
|
si:dkey-26m3.1
|
si:dkey-26m3.1 |
| chr6_-_18393476 | 0.58 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr22_-_16180467 | 0.58 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr4_-_49952636 | 0.57 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr20_-_54508650 | 0.56 |
ENSDART00000147642
|
CABZ01118770.1
|
|
| chr15_-_2010926 | 0.55 |
ENSDART00000155688
|
dock10
|
dedicator of cytokinesis 10 |
| chr14_+_6953244 | 0.55 |
ENSDART00000159470
|
rack1
|
receptor for activated C kinase 1 |
| chr7_+_26029672 | 0.53 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr2_+_44426609 | 0.53 |
ENSDART00000112711
ENSDART00000154188 |
kcnab1b
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 b |
| chr15_-_32383529 | 0.53 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr1_-_7582859 | 0.51 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr9_-_49860756 | 0.50 |
ENSDART00000044270
|
ttc21b
|
tetratricopeptide repeat domain 21B |
| chr10_-_43568239 | 0.50 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr25_+_31323978 | 0.49 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr24_+_39576071 | 0.49 |
ENSDART00000147137
|
si:dkey-161j23.6
|
si:dkey-161j23.6 |
| chr3_-_45281350 | 0.47 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr6_-_17266221 | 0.46 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr20_+_52458765 | 0.46 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr5_+_32141790 | 0.44 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr15_+_37412883 | 0.43 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr25_+_10793478 | 0.43 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr2_-_51794472 | 0.43 |
ENSDART00000186652
|
BX908782.3
|
|
| chr13_-_49819027 | 0.41 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr22_-_26005894 | 0.41 |
ENSDART00000105088
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr8_+_19548985 | 0.41 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr6_-_16804001 | 0.40 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr12_-_46959990 | 0.40 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr4_-_685412 | 0.39 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr6_-_16735402 | 0.39 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr19_+_7759354 | 0.39 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr7_+_17303944 | 0.39 |
ENSDART00000163505
ENSDART00000162169 ENSDART00000161291 |
nitr9
|
novel immune-type receptor 9 |
| chr19_+_25504645 | 0.37 |
ENSDART00000143292
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr21_-_27010796 | 0.37 |
ENSDART00000065398
ENSDART00000144342 ENSDART00000126542 |
ppp1r14ba
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ba |
| chr24_+_13635108 | 0.37 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr6_-_17052116 | 0.36 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr25_+_6186823 | 0.35 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr21_-_32284532 | 0.35 |
ENSDART00000190676
|
clk4b
|
CDC-like kinase 4b |
| chr4_+_11479705 | 0.34 |
ENSDART00000019458
ENSDART00000150587 ENSDART00000139370 ENSDART00000135826 |
asb13a.1
|
ankyrin repeat and SOCS box containing 13a, tandem duplicate 1 |
| chr4_+_34599803 | 0.33 |
ENSDART00000134088
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr1_-_9195629 | 0.32 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr4_-_57571688 | 0.31 |
ENSDART00000160179
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr24_+_13635387 | 0.29 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr5_-_42883761 | 0.29 |
ENSDART00000167374
|
BX323596.2
|
|
| chr15_-_32383340 | 0.29 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr22_-_26100282 | 0.28 |
ENSDART00000166075
|
pdgfaa
|
platelet-derived growth factor alpha polypeptide a |
| chr10_-_8434816 | 0.26 |
ENSDART00000108643
|
tctn1
|
tectonic family member 1 |
| chr2_-_14793343 | 0.25 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr8_-_45838481 | 0.24 |
ENSDART00000148206
ENSDART00000170485 |
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr17_-_26604549 | 0.24 |
ENSDART00000174773
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr18_-_49318823 | 0.24 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr16_+_34522977 | 0.23 |
ENSDART00000144069
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr8_-_28342487 | 0.22 |
ENSDART00000108508
|
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr2_-_49978227 | 0.21 |
ENSDART00000142835
|
hecw1b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1b |
| chr2_-_42035250 | 0.21 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr20_+_17581881 | 0.21 |
ENSDART00000182832
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr1_+_230363 | 0.20 |
ENSDART00000153285
|
tfdp1b
|
transcription factor Dp-1, b |
| chr15_-_3282220 | 0.20 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr24_+_25032340 | 0.20 |
ENSDART00000005845
|
mtmr6
|
myotubularin related protein 6 |
| chr25_-_30394746 | 0.19 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr1_-_10647484 | 0.19 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr15_+_29088426 | 0.18 |
ENSDART00000187290
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr16_-_41787421 | 0.17 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr18_+_17660402 | 0.15 |
ENSDART00000143475
|
cpne2
|
copine II |
| chr6_-_43677125 | 0.15 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr9_+_219124 | 0.14 |
ENSDART00000161484
|
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr1_-_10647307 | 0.13 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr11_-_34518362 | 0.13 |
ENSDART00000188223
|
pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr17_-_23631400 | 0.13 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr16_+_10841163 | 0.11 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr10_-_24868581 | 0.11 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr17_-_6536305 | 0.10 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr19_-_32804535 | 0.10 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr4_+_57580303 | 0.10 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr5_+_29160324 | 0.09 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr9_+_1313418 | 0.09 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr5_-_42071505 | 0.08 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr2_+_7192966 | 0.08 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr18_+_6479963 | 0.08 |
ENSDART00000092752
ENSDART00000136333 |
wash1
|
WAS protein family homolog 1 |
| chr3_-_46818001 | 0.07 |
ENSDART00000166505
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr12_-_30338779 | 0.07 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr20_+_37294112 | 0.07 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr4_+_49322310 | 0.07 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr17_-_43517542 | 0.06 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr3_-_46817838 | 0.05 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr1_+_33322555 | 0.04 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
| chr9_+_18572970 | 0.03 |
ENSDART00000114860
|
lacc1
|
laccase (multicopper oxidoreductase) domain containing 1 |
| chr20_+_46897504 | 0.02 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr15_-_31401361 | 0.02 |
ENSDART00000124360
|
or111-7
|
odorant receptor, family D, subfamily 111, member 7 |
| chr24_-_18477672 | 0.02 |
ENSDART00000126928
ENSDART00000158393 |
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr25_-_27842654 | 0.01 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr7_+_17106160 | 0.00 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr6_+_39923052 | 0.00 |
ENSDART00000149019
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat5a+stat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 0.7 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.2 | 0.6 | GO:0039529 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.2 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.2 | 0.9 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 0.5 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.1 | 0.7 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.1 | 0.5 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.8 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 2.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0090247 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.1 | 0.2 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 0.3 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 1.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 21.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.1 | 2.8 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.5 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.5 | GO:0043651 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.0 | 0.4 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.5 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 1.1 | GO:0042737 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.6 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.2 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.8 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.2 | GO:0001556 | oocyte maturation(GO:0001556) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 2.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.2 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.1 | 0.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.4 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.8 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.6 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.5 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.7 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 22.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.2 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.3 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.4 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |