Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
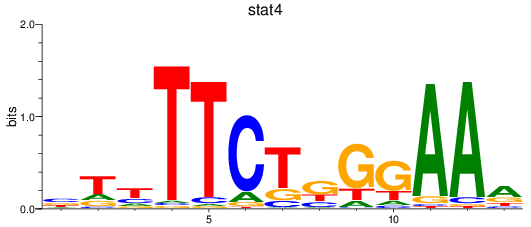
Results for stat4
Z-value: 0.60
Transcription factors associated with stat4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat4
|
ENSDARG00000028731 | signal transducer and activator of transcription 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat4 | dr11_v1_chr9_+_41156287_41156287 | -0.27 | 2.8e-01 | Click! |
Activity profile of stat4 motif
Sorted Z-values of stat4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_21005725 | 2.24 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr21_+_25777425 | 2.19 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr9_+_44994214 | 1.57 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr6_+_41503854 | 1.51 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr15_+_25489406 | 1.49 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr17_+_30591287 | 1.41 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr7_-_26518086 | 1.28 |
ENSDART00000058913
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr8_-_25033681 | 1.23 |
ENSDART00000003493
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr1_-_38171648 | 1.22 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr3_-_31879201 | 1.21 |
ENSDART00000076189
|
zgc:171779
|
zgc:171779 |
| chr21_-_32060993 | 1.18 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr1_-_6085750 | 1.16 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr17_-_12498096 | 1.15 |
ENSDART00000149551
ENSDART00000105215 ENSDART00000191207 |
emilin1b
|
elastin microfibril interfacer 1b |
| chr5_-_57624425 | 1.11 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr15_+_20529197 | 1.07 |
ENSDART00000060935
ENSDART00000137926 ENSDART00000140087 |
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr21_+_39100289 | 1.03 |
ENSDART00000075958
|
slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr9_-_55946377 | 1.02 |
ENSDART00000161536
ENSDART00000169432 |
SH3RF3
|
si:ch211-124n19.2 |
| chr14_-_33278084 | 1.01 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr18_-_3527988 | 1.00 |
ENSDART00000157669
|
capn5a
|
calpain 5a |
| chr1_-_36770883 | 0.99 |
ENSDART00000167831
|
prmt9
|
protein arginine methyltransferase 9 |
| chr7_+_32693890 | 0.98 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr3_+_36646054 | 0.98 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr18_-_3527686 | 0.97 |
ENSDART00000169049
|
capn5a
|
calpain 5a |
| chr18_-_20444296 | 0.96 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr7_-_50764714 | 0.93 |
ENSDART00000110283
|
iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr10_+_29770120 | 0.92 |
ENSDART00000100032
ENSDART00000193205 |
hyou1
|
hypoxia up-regulated 1 |
| chr6_-_8377055 | 0.90 |
ENSDART00000131513
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr1_+_12177195 | 0.87 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr19_+_43885770 | 0.87 |
ENSDART00000135599
|
lypla2
|
lysophospholipase II |
| chr12_+_28888975 | 0.85 |
ENSDART00000076362
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr4_+_16827390 | 0.83 |
ENSDART00000113344
|
kiss2
|
kisspeptin 2 |
| chr25_-_36263115 | 0.83 |
ENSDART00000143046
ENSDART00000139002 |
dus2
|
dihydrouridine synthase 2 |
| chr12_-_37299646 | 0.81 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr7_-_26497947 | 0.81 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr24_-_26820698 | 0.81 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr3_-_3439150 | 0.79 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr7_+_45963767 | 0.79 |
ENSDART00000163098
|
pop4
|
POP4 homolog, ribonuclease P/MRP subunit |
| chr1_-_24458057 | 0.78 |
ENSDART00000140197
ENSDART00000181286 |
sh3d19
|
SH3 domain containing 19 |
| chr12_-_35393211 | 0.72 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr6_-_11800427 | 0.70 |
ENSDART00000126243
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr5_+_67835595 | 0.69 |
ENSDART00000159386
|
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr7_+_34487833 | 0.69 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr23_-_33558161 | 0.68 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr22_-_11078988 | 0.68 |
ENSDART00000126664
ENSDART00000006927 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr19_+_38168006 | 0.68 |
ENSDART00000087662
ENSDART00000177759 |
phf14
|
PHD finger protein 14 |
| chr2_+_2957515 | 0.67 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr23_+_4350897 | 0.67 |
ENSDART00000190593
|
mybl2a
|
v-myb avian myeloblastosis viral oncogene homolog-like 2a |
| chr19_+_1673599 | 0.66 |
ENSDART00000163127
|
klhl7
|
kelch-like family member 7 |
| chr14_+_28473173 | 0.66 |
ENSDART00000017075
|
xiap
|
X-linked inhibitor of apoptosis |
| chr19_-_3821678 | 0.64 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr16_+_16266428 | 0.63 |
ENSDART00000188433
|
setd2
|
SET domain containing 2 |
| chr16_-_2390931 | 0.62 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr22_-_11493236 | 0.61 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr19_-_46091497 | 0.61 |
ENSDART00000178772
ENSDART00000167255 |
ptdss1b
si:dkey-108k24.2
|
phosphatidylserine synthase 1b si:dkey-108k24.2 |
| chr23_+_29966466 | 0.61 |
ENSDART00000143583
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr13_-_9300299 | 0.59 |
ENSDART00000144142
|
HTRA2 (1 of many)
|
si:dkey-33c12.12 |
| chr16_-_22585289 | 0.58 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr19_-_43750389 | 0.58 |
ENSDART00000147328
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr7_+_38770167 | 0.57 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr11_+_19603251 | 0.56 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr16_-_9830451 | 0.55 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr7_+_29065915 | 0.54 |
ENSDART00000136657
|
vrk3
|
vaccinia related kinase 3 |
| chr9_-_27738110 | 0.54 |
ENSDART00000060347
|
crygs2
|
crystallin, gamma S2 |
| chr6_+_2195625 | 0.52 |
ENSDART00000155659
|
acvr1bb
|
activin A receptor type 1Bb |
| chr14_-_32631519 | 0.52 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase phospholipid transporting 11C |
| chr7_-_24875421 | 0.52 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr20_-_30938184 | 0.52 |
ENSDART00000147359
ENSDART00000062552 |
wtap
|
WT1 associated protein |
| chr22_+_20172018 | 0.52 |
ENSDART00000188104
|
hmg20b
|
high mobility group 20B |
| chr2_+_16449627 | 0.51 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr12_+_33403694 | 0.51 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr23_+_43718115 | 0.51 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr13_+_8255106 | 0.50 |
ENSDART00000080465
|
hells
|
helicase, lymphoid specific |
| chr21_+_20488662 | 0.49 |
ENSDART00000147951
|
efna5a
|
ephrin-A5a |
| chr15_-_23721618 | 0.49 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr5_-_37959874 | 0.47 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr5_+_36768674 | 0.47 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr10_+_8401929 | 0.47 |
ENSDART00000059028
|
hvcn1
|
hydrogen voltage-gated channel 1 |
| chr3_-_32275975 | 0.47 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr1_-_493218 | 0.47 |
ENSDART00000031635
|
ercc5
|
excision repair cross-complementation group 5 |
| chr21_-_11970199 | 0.44 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr21_-_40049642 | 0.43 |
ENSDART00000124416
|
med31
|
mediator complex subunit 31 |
| chr7_-_29043075 | 0.43 |
ENSDART00000052342
|
thap11
|
THAP domain containing 11 |
| chr8_-_18203274 | 0.43 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr3_+_40289418 | 0.43 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr5_-_41984298 | 0.43 |
ENSDART00000189535
|
ncor1
|
nuclear receptor corepressor 1 |
| chr2_-_9989919 | 0.42 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr6_+_29386728 | 0.42 |
ENSDART00000104303
|
actl6a
|
actin-like 6A |
| chr25_+_16312258 | 0.42 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr5_+_54280135 | 0.42 |
ENSDART00000170218
ENSDART00000166050 |
rangrf
|
RAN guanine nucleotide release factor |
| chr6_-_3998199 | 0.42 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr12_+_36109507 | 0.42 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr9_+_38502524 | 0.41 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr23_+_1216215 | 0.41 |
ENSDART00000165957
|
utrn
|
utrophin |
| chr13_-_36049008 | 0.40 |
ENSDART00000189978
|
lgmn
|
legumain |
| chr21_+_20901505 | 0.40 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr2_+_42005217 | 0.40 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr21_-_3613702 | 0.40 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr18_+_7543347 | 0.39 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr8_-_18203092 | 0.38 |
ENSDART00000140620
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr25_+_32390794 | 0.38 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr23_-_5783421 | 0.36 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr25_+_11281970 | 0.35 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr22_-_20838011 | 0.35 |
ENSDART00000186056
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr14_-_22015232 | 0.35 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr6_+_4387150 | 0.35 |
ENSDART00000181283
|
rbm26
|
RNA binding motif protein 26 |
| chr23_-_43718067 | 0.34 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr3_-_61377127 | 0.34 |
ENSDART00000155932
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr19_-_12315693 | 0.34 |
ENSDART00000151158
|
ncaldb
|
neurocalcin delta b |
| chr22_-_28645676 | 0.33 |
ENSDART00000005883
|
jagn1b
|
jagunal homolog 1b |
| chr3_-_13599482 | 0.33 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr21_+_39197628 | 0.33 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr23_-_17451264 | 0.32 |
ENSDART00000190748
|
tpd52l2b
|
tumor protein D52-like 2b |
| chr16_-_4610255 | 0.32 |
ENSDART00000081852
ENSDART00000123253 ENSDART00000127554 ENSDART00000029485 |
arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr22_-_20514043 | 0.32 |
ENSDART00000088890
|
atp8b3
|
ATPase phospholipid transporting 8B3 |
| chr3_-_43954971 | 0.32 |
ENSDART00000167159
ENSDART00000189315 |
ubfd1
|
ubiquitin family domain containing 1 |
| chr2_+_42005475 | 0.32 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr6_-_30689126 | 0.31 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial |
| chr8_+_47897734 | 0.31 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr18_-_21047580 | 0.31 |
ENSDART00000010189
|
igf1ra
|
insulin-like growth factor 1a receptor |
| chr5_+_36895545 | 0.30 |
ENSDART00000135776
ENSDART00000147561 ENSDART00000133842 ENSDART00000051185 ENSDART00000141984 ENSDART00000136301 ENSDART00000142388 |
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr19_+_40379771 | 0.30 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr24_+_37484661 | 0.30 |
ENSDART00000165125
ENSDART00000109221 |
wdr90
|
WD repeat domain 90 |
| chr10_-_34033680 | 0.30 |
ENSDART00000180734
|
rfc3
|
replication factor C (activator 1) 3 |
| chr2_-_16449504 | 0.29 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr1_-_18585046 | 0.29 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr8_-_11067079 | 0.29 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr22_-_9861531 | 0.29 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr8_-_20838342 | 0.28 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr12_+_23371542 | 0.28 |
ENSDART00000078896
|
waca
|
WW domain containing adaptor with coiled-coil a |
| chr11_+_5681762 | 0.27 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr11_+_41459408 | 0.27 |
ENSDART00000182285
|
park7
|
parkinson protein 7 |
| chr12_-_26383242 | 0.26 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr7_-_34434889 | 0.25 |
ENSDART00000159982
|
nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr2_+_37838259 | 0.25 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr22_-_22130623 | 0.25 |
ENSDART00000113168
|
CU855878.1
|
|
| chr16_+_45746549 | 0.25 |
ENSDART00000190403
|
paqr6
|
progestin and adipoQ receptor family member VI |
| chr14_-_45595711 | 0.24 |
ENSDART00000074038
|
scyl1
|
SCY1-like, kinase-like 1 |
| chr23_-_31763753 | 0.24 |
ENSDART00000053399
|
aldh8a1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr14_+_36521005 | 0.24 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr17_+_24111392 | 0.23 |
ENSDART00000180123
ENSDART00000182787 ENSDART00000189752 ENSDART00000184940 ENSDART00000185363 ENSDART00000064067 |
ehbp1
|
EH domain binding protein 1 |
| chr15_-_26538989 | 0.23 |
ENSDART00000032880
|
rpa1
|
replication protein A1 |
| chr19_-_22843480 | 0.23 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr15_-_31508221 | 0.22 |
ENSDART00000121464
|
si:dkey-1m11.5
|
si:dkey-1m11.5 |
| chr24_-_20599781 | 0.22 |
ENSDART00000179664
ENSDART00000141823 |
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr19_-_10286228 | 0.22 |
ENSDART00000027316
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr8_+_54055390 | 0.21 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr5_+_36895860 | 0.20 |
ENSDART00000134493
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr11_+_43401201 | 0.20 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr20_-_28433616 | 0.20 |
ENSDART00000169289
|
wdr21
|
WD repeat domain 21 |
| chr14_-_52433292 | 0.20 |
ENSDART00000164272
|
rest
|
RE1-silencing transcription factor |
| chr19_-_17658160 | 0.20 |
ENSDART00000151766
ENSDART00000170790 ENSDART00000186678 ENSDART00000188045 ENSDART00000176980 ENSDART00000166313 ENSDART00000188589 |
thrb
|
thyroid hormone receptor beta |
| chr23_-_25779995 | 0.20 |
ENSDART00000110670
|
si:dkey-21c19.3
|
si:dkey-21c19.3 |
| chr17_+_12767640 | 0.20 |
ENSDART00000112469
ENSDART00000154984 ENSDART00000124584 ENSDART00000193647 ENSDART00000179686 |
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr1_-_28831848 | 0.18 |
ENSDART00000148536
|
GK3P
|
zgc:172295 |
| chr9_-_16062938 | 0.18 |
ENSDART00000099479
|
crfb4
|
cytokine receptor family member b4 |
| chr4_-_1818315 | 0.18 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr23_-_24483311 | 0.18 |
ENSDART00000185793
ENSDART00000109248 |
spen
|
spen family transcriptional repressor |
| chr19_-_2317558 | 0.17 |
ENSDART00000190300
|
sp8a
|
sp8 transcription factor a |
| chr6_+_18367388 | 0.17 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr2_+_25868404 | 0.17 |
ENSDART00000143517
ENSDART00000187743 |
slc7a14a
|
solute carrier family 7, member 14a |
| chr17_-_45040813 | 0.17 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr14_+_45970033 | 0.16 |
ENSDART00000047716
|
fermt3b
|
fermitin family member 3b |
| chr18_+_17725410 | 0.16 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr9_+_27354653 | 0.15 |
ENSDART00000134134
|
tlr20.2
|
toll-like receptor 20, tandem duplicate 2 |
| chr17_+_32571584 | 0.15 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr17_-_8173380 | 0.15 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr19_+_7938121 | 0.14 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr11_-_11910225 | 0.14 |
ENSDART00000159922
|
si:ch211-69b7.6
|
si:ch211-69b7.6 |
| chr1_+_37391716 | 0.14 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr12_-_9796237 | 0.14 |
ENSDART00000149344
|
prdm9
|
PR domain containing 9 |
| chr20_+_18994059 | 0.13 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr10_+_20392083 | 0.13 |
ENSDART00000166867
|
r3hcc1
|
R3H domain and coiled-coil containing 1 |
| chr15_-_31390985 | 0.13 |
ENSDART00000060117
|
or111-6
|
odorant receptor, family D, subfamily 111, member 6 |
| chr6_+_18423402 | 0.12 |
ENSDART00000159747
|
rab11fip4b
|
RAB11 family interacting protein 4 (class II) b |
| chr5_-_40190949 | 0.12 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr7_-_27696958 | 0.11 |
ENSDART00000173470
|
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr17_-_45386546 | 0.11 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr10_-_36207737 | 0.11 |
ENSDART00000159366
|
or109-13
|
odorant receptor, family D, subfamily 109, member 13 |
| chr13_+_12405051 | 0.11 |
ENSDART00000108543
|
atp10d
|
ATPase phospholipid transporting 10D |
| chr19_-_43750108 | 0.11 |
ENSDART00000171806
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr15_-_31390760 | 0.10 |
ENSDART00000190774
|
or111-6
|
odorant receptor, family D, subfamily 111, member 6 |
| chr12_+_30367079 | 0.10 |
ENSDART00000190112
|
ccdc186
|
si:ch211-225b10.4 |
| chr20_+_10166297 | 0.10 |
ENSDART00000141877
|
kcnk10a
|
potassium channel, subfamily K, member 10a |
| chr16_+_38394371 | 0.10 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr17_-_2834764 | 0.10 |
ENSDART00000024027
|
bdkrb1
|
bradykinin receptor B1 |
| chr11_-_11882982 | 0.10 |
ENSDART00000190853
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr4_-_13518381 | 0.10 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr11_+_6431133 | 0.10 |
ENSDART00000190742
|
FO904935.1
|
|
| chr21_+_1378250 | 0.10 |
ENSDART00000186912
|
TCF4
|
transcription factor 4 |
| chr20_-_43775495 | 0.10 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr3_+_3852614 | 0.09 |
ENSDART00000184334
ENSDART00000162313 |
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr24_+_18299506 | 0.09 |
ENSDART00000172056
|
tpk1
|
thiamin pyrophosphokinase 1 |
| chr16_-_11859309 | 0.09 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr21_+_39197418 | 0.09 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr15_-_5245726 | 0.09 |
ENSDART00000174079
ENSDART00000174041 |
or128-1
|
odorant receptor, family E, subfamily 128, member 1 |
| chr1_-_53407087 | 0.09 |
ENSDART00000157910
|
elmod2
|
ELMO/CED-12 domain containing 2 |
| chr12_-_17602958 | 0.09 |
ENSDART00000134690
ENSDART00000028090 |
eif2ak1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr16_-_10071886 | 0.08 |
ENSDART00000136168
ENSDART00000130382 |
ptpn2a
|
protein tyrosine phosphatase, non-receptor type 2, a |
| chr3_+_58119571 | 0.08 |
ENSDART00000108979
|
si:ch211-256e16.4
|
si:ch211-256e16.4 |
| chr3_-_31057624 | 0.08 |
ENSDART00000152901
|
armc5
|
armadillo repeat containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.3 | 1.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.3 | 0.9 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.2 | 1.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.2 | 0.9 | GO:0071938 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.2 | 0.6 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.2 | 0.5 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 1.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.8 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.1 | 1.6 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.7 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.3 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.3 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.4 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.3 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.1 | 0.5 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 0.6 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.3 | GO:1904729 | regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.1 | 1.1 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 0.5 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.1 | 0.2 | GO:1903430 | branchiomotor neuron axon guidance(GO:0021785) negative regulation of cell maturation(GO:1903430) |
| 0.1 | 0.4 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.1 | 0.6 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.2 | GO:0042182 | ketone catabolic process(GO:0042182) |
| 0.1 | 0.3 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.1 | 1.0 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.1 | 0.5 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.2 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.7 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.0 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0042671 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.4 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.5 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:0044783 | G1 DNA damage checkpoint(GO:0044783) |
| 0.0 | 0.5 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.3 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.1 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.9 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.0 | 0.2 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.3 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.1 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.0 | 0.3 | GO:0033753 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.8 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.7 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.5 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.6 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.7 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.7 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.1 | GO:0050996 | positive regulation of lipid catabolic process(GO:0050996) |
| 0.0 | 0.6 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.4 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0043967 | histone H4 acetylation(GO:0043967) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 1.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 1.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.2 | 0.5 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.4 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.5 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.8 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.1 | 1.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 0.3 | GO:1990745 | EARP complex(GO:1990745) |
| 0.1 | 0.6 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.4 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 1.1 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 2.2 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.7 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.6 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.2 | 0.9 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.8 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 1.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.1 | 1.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.1 | 0.3 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.0 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 1.6 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 2.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.4 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.2 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.8 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.8 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.2 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.7 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.4 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 2.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.3 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.2 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.9 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 1.0 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.4 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.4 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.3 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.8 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |