Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
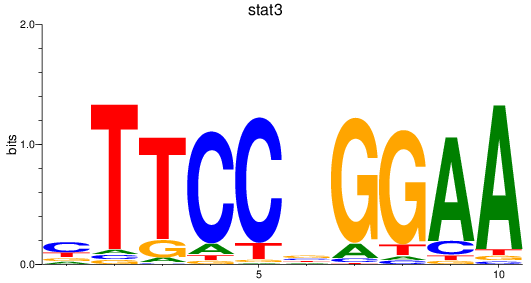
Results for stat3
Z-value: 0.25
Transcription factors associated with stat3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat3
|
ENSDARG00000022712 | signal transducer and activator of transcription 3 (acute-phase response factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat3 | dr11_v1_chr3_+_17030665_17030704 | 0.06 | 8.3e-01 | Click! |
Activity profile of stat3 motif
Sorted Z-values of stat3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_17964525 | 0.95 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr24_+_31361407 | 0.75 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr1_-_51734524 | 0.63 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr6_-_40352215 | 0.63 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr3_+_39922602 | 0.61 |
ENSDART00000193937
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr24_-_37568359 | 0.59 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr22_+_36914636 | 0.57 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr13_-_37608441 | 0.52 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr21_+_35215810 | 0.50 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr19_+_17385561 | 0.48 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr18_-_21177674 | 0.47 |
ENSDART00000060175
|
si:dkey-12e7.4
|
si:dkey-12e7.4 |
| chr13_-_37642890 | 0.47 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr9_+_48219111 | 0.47 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr14_-_30747686 | 0.47 |
ENSDART00000008373
|
fosl1a
|
FOS-like antigen 1a |
| chr1_-_9114936 | 0.47 |
ENSDART00000163624
ENSDART00000132910 |
si:ch211-14k19.8
vap
|
si:ch211-14k19.8 vascular associated protein |
| chr9_+_40825065 | 0.44 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr3_-_35541378 | 0.43 |
ENSDART00000183075
ENSDART00000022147 |
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr23_-_19715799 | 0.41 |
ENSDART00000142072
ENSDART00000032744 ENSDART00000131860 |
rpl10
|
ribosomal protein L10 |
| chr7_+_32369026 | 0.40 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr19_+_712127 | 0.39 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr8_-_7502166 | 0.38 |
ENSDART00000176938
|
cdk20
|
cyclin-dependent kinase 20 |
| chr3_-_31655796 | 0.38 |
ENSDART00000181052
|
CU062431.1
|
|
| chr14_+_11395383 | 0.38 |
ENSDART00000106657
|
glod5
|
glyoxalase domain containing 5 |
| chr16_-_35394422 | 0.38 |
ENSDART00000180593
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr17_+_40989973 | 0.37 |
ENSDART00000160049
|
mrpl33
|
mitochondrial ribosomal protein L33 |
| chr16_-_12809873 | 0.37 |
ENSDART00000146997
ENSDART00000178291 ENSDART00000007842 |
isoc2
|
isochorismatase domain containing 2 |
| chr21_-_30545121 | 0.36 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr3_-_22829710 | 0.35 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr3_+_26734162 | 0.35 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr16_+_40560622 | 0.34 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr4_-_18436899 | 0.34 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr1_-_46981134 | 0.34 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr3_-_57425961 | 0.33 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr22_+_7480465 | 0.33 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr20_-_35841570 | 0.33 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr6_+_43015916 | 0.31 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr3_+_59899452 | 0.31 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr21_+_8341774 | 0.30 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr18_+_3332999 | 0.29 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr3_-_13461056 | 0.28 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr17_+_7522777 | 0.28 |
ENSDART00000184723
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr16_+_54513267 | 0.27 |
ENSDART00000161923
|
CABZ01088508.1
|
|
| chr10_-_3332362 | 0.27 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr14_-_28567845 | 0.26 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr19_+_5418006 | 0.26 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr6_+_33897624 | 0.26 |
ENSDART00000166706
|
ccdc17
|
coiled-coil domain containing 17 |
| chr16_+_31922065 | 0.26 |
ENSDART00000131661
ENSDART00000144194 ENSDART00000145510 |
rps9
|
ribosomal protein S9 |
| chr1_-_26398361 | 0.26 |
ENSDART00000160183
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr11_-_6265574 | 0.26 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr3_+_472158 | 0.25 |
ENSDART00000134971
|
zgc:194659
|
zgc:194659 |
| chr19_+_47783137 | 0.25 |
ENSDART00000024777
ENSDART00000158979 |
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr23_-_40792128 | 0.25 |
ENSDART00000145360
|
si:dkey-194e6.2
|
si:dkey-194e6.2 |
| chr13_+_6092135 | 0.24 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr16_+_31921812 | 0.24 |
ENSDART00000176928
ENSDART00000193733 |
rps9
|
ribosomal protein S9 |
| chr24_+_5208171 | 0.24 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr9_+_26103814 | 0.23 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr13_-_49819027 | 0.23 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr15_-_29510074 | 0.22 |
ENSDART00000184070
|
BX324227.3
|
|
| chr12_-_19346678 | 0.22 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr24_-_37472727 | 0.22 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr17_+_25397070 | 0.21 |
ENSDART00000164254
|
zgc:154055
|
zgc:154055 |
| chr1_+_30083578 | 0.21 |
ENSDART00000101779
ENSDART00000143639 |
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr20_+_52458765 | 0.21 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr3_-_13461361 | 0.21 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr14_+_11457500 | 0.20 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr7_+_32369463 | 0.20 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr1_-_10071422 | 0.20 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr25_+_34246625 | 0.20 |
ENSDART00000082578
|
bnip2
|
BCL2 interacting protein 2 |
| chr5_+_15819651 | 0.19 |
ENSDART00000081230
ENSDART00000186969 ENSDART00000134206 |
hspb8
|
heat shock protein b8 |
| chr4_-_12323228 | 0.19 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr6_+_22679610 | 0.18 |
ENSDART00000102701
|
zmp:0000000634
|
zmp:0000000634 |
| chr20_-_47188966 | 0.18 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr3_+_4502066 | 0.18 |
ENSDART00000088610
|
rangap1a
|
RAN GTPase activating protein 1a |
| chr1_-_17711636 | 0.18 |
ENSDART00000148322
ENSDART00000122670 |
ufsp2
|
ufm1-specific peptidase 2 |
| chr24_+_16149514 | 0.18 |
ENSDART00000152087
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr24_+_16149251 | 0.18 |
ENSDART00000188289
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr2_+_26288301 | 0.17 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr22_-_15578402 | 0.17 |
ENSDART00000062986
|
hsh2d
|
hematopoietic SH2 domain containing |
| chr22_-_834106 | 0.17 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr22_+_20427170 | 0.17 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr20_-_23440955 | 0.17 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr10_-_4375190 | 0.17 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr2_+_17181777 | 0.17 |
ENSDART00000112063
|
ptger4c
|
prostaglandin E receptor 4 (subtype EP4) c |
| chr19_+_43359075 | 0.16 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr4_+_359970 | 0.16 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr21_+_45626136 | 0.16 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr17_+_10318071 | 0.16 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr7_-_22941472 | 0.16 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr21_+_13150937 | 0.16 |
ENSDART00000102251
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr21_-_5879897 | 0.16 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr1_+_5402476 | 0.16 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr12_-_32013125 | 0.15 |
ENSDART00000153355
|
grin2cb
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2Cb |
| chr19_-_41405187 | 0.15 |
ENSDART00000038038
ENSDART00000172350 |
sem1
|
SEM1, 26S proteasome complex subunit |
| chr8_+_20918207 | 0.15 |
ENSDART00000144039
|
si:ch73-196i15.5
|
si:ch73-196i15.5 |
| chr4_+_76735113 | 0.15 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr18_+_37219140 | 0.15 |
ENSDART00000088247
|
qpctla
|
glutaminyl-peptide cyclotransferase-like a |
| chr14_+_15257658 | 0.15 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr22_-_16180467 | 0.15 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr18_-_3166726 | 0.14 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr21_-_27338639 | 0.14 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr22_-_372723 | 0.14 |
ENSDART00000112895
|
FNDC10
|
si:zfos-1324h11.5 |
| chr19_+_10587307 | 0.14 |
ENSDART00000131225
|
si:dkey-211g8.4
|
si:dkey-211g8.4 |
| chr20_+_53521648 | 0.14 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr14_+_6159356 | 0.14 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr3_+_1219344 | 0.13 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr3_-_44059902 | 0.13 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr4_+_47257854 | 0.13 |
ENSDART00000173868
|
crestin
|
crestin |
| chr10_-_20105553 | 0.13 |
ENSDART00000184502
|
pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr22_-_12890921 | 0.13 |
ENSDART00000146484
|
nab1a
|
NGFI-A binding protein 1a (EGR1 binding protein 1) |
| chr3_-_40933415 | 0.13 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr5_+_32009956 | 0.13 |
ENSDART00000188482
|
scai
|
suppressor of cancer cell invasion |
| chr21_-_21781158 | 0.13 |
ENSDART00000113734
|
chrdl2
|
chordin-like 2 |
| chr3_-_6709938 | 0.13 |
ENSDART00000172196
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr9_-_23922011 | 0.13 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr11_-_762721 | 0.13 |
ENSDART00000166465
|
syn2b
|
synapsin IIb |
| chr7_+_25053331 | 0.13 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr24_-_27461295 | 0.12 |
ENSDART00000110748
|
ccl34b.9
|
chemokine (C-C motif) ligand 34b, duplicate 9 |
| chr5_+_24179307 | 0.12 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr1_-_9195629 | 0.12 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr24_-_39858710 | 0.12 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
| chr16_-_31717851 | 0.12 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr20_-_39273987 | 0.12 |
ENSDART00000127173
|
clu
|
clusterin |
| chr24_+_37406535 | 0.12 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr17_-_31308658 | 0.12 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr24_-_26485098 | 0.12 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr7_-_28696556 | 0.12 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr22_+_1462177 | 0.12 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr7_+_41313568 | 0.11 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr6_+_11989537 | 0.11 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr3_-_44012748 | 0.11 |
ENSDART00000167248
ENSDART00000157463 ENSDART00000159111 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr18_-_16801033 | 0.11 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr18_-_14860435 | 0.11 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr12_+_17042754 | 0.11 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr2_+_4402765 | 0.11 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr13_-_50366950 | 0.11 |
ENSDART00000098209
|
sirt1
|
sirtuin 1 |
| chr18_-_28938912 | 0.11 |
ENSDART00000136201
|
si:ch211-174j14.2
|
si:ch211-174j14.2 |
| chr22_+_3238474 | 0.11 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr24_-_33291784 | 0.10 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr16_-_19254323 | 0.10 |
ENSDART00000138744
|
dnah11
|
dynein, axonemal, heavy chain 11 |
| chr5_-_40734045 | 0.10 |
ENSDART00000010896
|
isl1
|
ISL LIM homeobox 1 |
| chr16_-_29458806 | 0.10 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr21_+_19925910 | 0.10 |
ENSDART00000111694
ENSDART00000132653 |
tnksa
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase a |
| chr19_+_7759354 | 0.10 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr20_+_32497260 | 0.10 |
ENSDART00000145175
ENSDART00000132921 |
si:ch73-257c13.2
|
si:ch73-257c13.2 |
| chr24_-_26383978 | 0.10 |
ENSDART00000031426
|
skilb
|
SKI-like proto-oncogene b |
| chr9_+_13985567 | 0.10 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr14_+_6159162 | 0.10 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr6_-_35106425 | 0.10 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr14_-_49859747 | 0.10 |
ENSDART00000169456
ENSDART00000164967 |
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr14_+_7452302 | 0.10 |
ENSDART00000130388
|
gfra3
|
GDNF family receptor alpha 3 |
| chr24_+_13635108 | 0.10 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr1_+_51827046 | 0.09 |
ENSDART00000052992
|
dand5
|
DAN domain family, member 5 |
| chr19_-_26863626 | 0.09 |
ENSDART00000145568
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr5_-_49012569 | 0.09 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr15_+_23550890 | 0.09 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr2_+_21063660 | 0.09 |
ENSDART00000022765
|
riok1
|
RIO kinase 1 (yeast) |
| chr21_-_4695583 | 0.09 |
ENSDART00000031425
|
zgc:55582
|
zgc:55582 |
| chr16_-_1502699 | 0.09 |
ENSDART00000187189
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr24_+_13635387 | 0.09 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr16_-_31718013 | 0.09 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr20_+_25552057 | 0.09 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr3_-_32170850 | 0.09 |
ENSDART00000055307
ENSDART00000157366 |
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr4_+_76637548 | 0.08 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr24_-_37877743 | 0.08 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr9_+_29585943 | 0.08 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr11_-_287670 | 0.08 |
ENSDART00000035737
|
slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr8_+_52491436 | 0.08 |
ENSDART00000164298
|
si:ch1073-392o20.1
|
si:ch1073-392o20.1 |
| chr16_-_27749172 | 0.08 |
ENSDART00000145198
|
steap4
|
STEAP family member 4 |
| chr17_-_23603263 | 0.08 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr6_+_40573365 | 0.08 |
ENSDART00000004075
|
uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr21_-_34844059 | 0.08 |
ENSDART00000136402
|
zgc:56585
|
zgc:56585 |
| chr1_-_10647484 | 0.08 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr11_+_11200550 | 0.08 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr3_-_16039619 | 0.08 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr25_-_28587896 | 0.08 |
ENSDART00000111908
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr19_+_8973042 | 0.07 |
ENSDART00000039597
|
crabp2b
|
cellular retinoic acid binding protein 2, b |
| chr7_+_58686860 | 0.07 |
ENSDART00000052332
|
penkb
|
proenkephalin b |
| chr5_-_42883761 | 0.07 |
ENSDART00000167374
|
BX323596.2
|
|
| chr2_-_37837056 | 0.07 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr4_+_29206813 | 0.07 |
ENSDART00000131893
|
si:dkey-23a23.1
|
si:dkey-23a23.1 |
| chr10_+_22134606 | 0.07 |
ENSDART00000155228
|
si:dkey-4c2.11
|
si:dkey-4c2.11 |
| chr7_+_48319916 | 0.07 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr11_-_29996344 | 0.07 |
ENSDART00000003712
ENSDART00000126110 |
ace2
|
angiotensin I converting enzyme 2 |
| chr9_-_23922778 | 0.07 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr16_+_32059785 | 0.07 |
ENSDART00000134459
|
si:dkey-40m6.8
|
si:dkey-40m6.8 |
| chr13_-_37631092 | 0.07 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr21_+_40443118 | 0.07 |
ENSDART00000190675
|
ssh2b
|
slingshot protein phosphatase 2b |
| chr21_+_20901505 | 0.06 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr17_+_53292215 | 0.06 |
ENSDART00000170686
|
si:ch1073-416d2.3
|
si:ch1073-416d2.3 |
| chr19_+_34311374 | 0.06 |
ENSDART00000086617
|
gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr7_+_27277236 | 0.06 |
ENSDART00000185161
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr2_+_10766744 | 0.06 |
ENSDART00000015379
|
gfi1aa
|
growth factor independent 1A transcription repressor a |
| chr18_-_44888375 | 0.06 |
ENSDART00000160506
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr18_+_10820430 | 0.06 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr6_+_55357188 | 0.06 |
ENSDART00000158219
|
pltp
|
phospholipid transfer protein |
| chr9_-_54304684 | 0.06 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr23_-_5101847 | 0.06 |
ENSDART00000122240
|
etv7
|
ets variant 7 |
| chr20_-_25412181 | 0.06 |
ENSDART00000062780
|
itsn2a
|
intersectin 2a |
| chr16_+_25809235 | 0.06 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr2_-_37837472 | 0.06 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.5 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.1 | 0.3 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.1 | 0.6 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.3 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.1 | 0.2 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.2 | GO:0050968 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.0 | 0.3 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.1 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.2 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0042546 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.1 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.5 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0002839 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.2 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.1 | GO:0045901 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.0 | 0.0 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.3 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0002831 | regulation of response to biotic stimulus(GO:0002831) regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.0 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.0 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.7 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.5 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.4 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.4 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.2 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.2 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.5 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 1.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |