Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
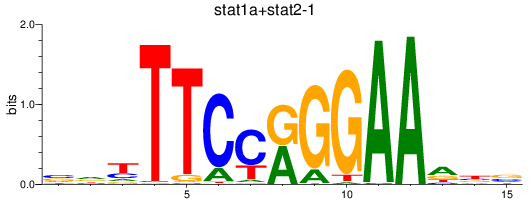
Results for stat1a+stat2-1_stat1b
Z-value: 0.67
Transcription factors associated with stat1a+stat2-1_stat1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat1a
|
ENSDARG00000006266 | signal transducer and activator of transcription 1a |
|
stat2-1
|
ENSDARG00000031647 | signal transducer and activator of transcription 2 |
|
stat1b
|
ENSDARG00000076182 | signal transducer and activator of transcription 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat1b | dr11_v1_chr9_+_41218967_41218974 | 0.95 | 2.0e-09 | Click! |
| stat2 | dr11_v1_chr6_-_39160422_39160422 | 0.94 | 9.2e-09 | Click! |
| stat1a | dr11_v1_chr22_+_12798569_12798569 | 0.59 | 9.6e-03 | Click! |
Activity profile of stat1a+stat2-1_stat1b motif
Sorted Z-values of stat1a+stat2-1_stat1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_17964525 | 3.76 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr24_+_31361407 | 3.63 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr6_-_40352215 | 3.34 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr9_+_40825065 | 2.69 |
ENSDART00000137673
|
si:dkey-95p16.2
|
si:dkey-95p16.2 |
| chr14_+_33329420 | 2.39 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr13_-_37608441 | 2.26 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr14_+_33329761 | 1.87 |
ENSDART00000161138
|
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr24_+_24923166 | 1.84 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr12_-_3133483 | 1.64 |
ENSDART00000015092
|
col1a1b
|
collagen, type I, alpha 1b |
| chr20_+_23440632 | 1.60 |
ENSDART00000180685
ENSDART00000042820 |
si:dkey-90m5.4
|
si:dkey-90m5.4 |
| chr7_+_32369026 | 1.58 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr14_-_30747686 | 1.51 |
ENSDART00000008373
|
fosl1a
|
FOS-like antigen 1a |
| chr8_+_26522013 | 1.38 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr19_+_5418006 | 1.38 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr22_+_7480465 | 1.28 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr19_+_17385561 | 1.27 |
ENSDART00000141397
ENSDART00000143913 ENSDART00000133626 |
rpl15
|
ribosomal protein L15 |
| chr8_+_40573137 | 1.24 |
ENSDART00000184278
|
izumo1
|
izumo sperm-egg fusion 1 |
| chr22_-_16180467 | 1.19 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr9_+_52411530 | 1.19 |
ENSDART00000163684
|
nme8
|
NME/NM23 family member 8 |
| chr21_+_45626136 | 1.18 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr2_+_26288301 | 1.18 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr6_-_8311044 | 1.17 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr24_+_5208171 | 1.15 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr10_-_4375190 | 1.15 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr10_-_3332362 | 1.09 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr4_-_13567387 | 1.05 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr8_-_47212686 | 1.00 |
ENSDART00000040370
|
pimr189
|
Pim proto-oncogene, serine/threonine kinase, related 189 |
| chr7_+_26029672 | 0.98 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr14_+_24241241 | 0.95 |
ENSDART00000022377
|
nkx2.5
|
NK2 homeobox 5 |
| chr21_-_27338639 | 0.94 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr8_-_46060685 | 0.93 |
ENSDART00000121902
|
pimr188
|
Pim proto-oncogene, serine/threonine kinase, related 188 |
| chr1_-_46981134 | 0.90 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr15_-_32383529 | 0.89 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr14_-_21123551 | 0.88 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr21_+_26720803 | 0.87 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr20_-_35841570 | 0.85 |
ENSDART00000037855
|
tnfrsf21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr13_+_33117528 | 0.85 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr3_+_39853788 | 0.84 |
ENSDART00000154869
|
cacna1ha
|
calcium channel, voltage-dependent, T type, alpha 1H subunit a |
| chr4_+_19127973 | 0.83 |
ENSDART00000136611
|
si:dkey-21o22.2
|
si:dkey-21o22.2 |
| chr15_-_25083200 | 0.83 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr15_-_25010400 | 0.83 |
ENSDART00000155236
|
si:ch211-198e20.10
|
si:ch211-198e20.10 |
| chr21_+_8341774 | 0.82 |
ENSDART00000129749
ENSDART00000055325 ENSDART00000133804 |
psmb7
|
proteasome subunit beta 7 |
| chr20_+_52458765 | 0.82 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr21_+_35215810 | 0.81 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr7_+_32369463 | 0.81 |
ENSDART00000180544
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr11_+_30321116 | 0.79 |
ENSDART00000187921
ENSDART00000127075 |
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr4_-_12323228 | 0.79 |
ENSDART00000081089
|
il17ra1a
|
interleukin 17 receptor A1a |
| chr24_-_38197040 | 0.78 |
ENSDART00000137949
ENSDART00000105639 |
igic1s1
|
immunoglobulin light iota constant 1, s1 |
| chr3_-_13461056 | 0.77 |
ENSDART00000137678
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr7_+_39706004 | 0.77 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr4_-_1497384 | 0.76 |
ENSDART00000093236
|
zmp:0000000711
|
zmp:0000000711 |
| chr11_-_27917730 | 0.76 |
ENSDART00000173219
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr24_+_16149514 | 0.75 |
ENSDART00000152087
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr16_+_40560622 | 0.73 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr24_+_16149251 | 0.72 |
ENSDART00000188289
|
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr15_-_25039083 | 0.72 |
ENSDART00000154409
|
pimr190
|
Pim proto-oncogene, serine/threonine kinase, related 190 |
| chr14_-_21123325 | 0.71 |
ENSDART00000180889
ENSDART00000188539 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr3_-_40933415 | 0.71 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr15_+_37412883 | 0.67 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr21_+_43404945 | 0.67 |
ENSDART00000142234
|
frmd7
|
FERM domain containing 7 |
| chr6_+_43015916 | 0.66 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr3_-_13461361 | 0.65 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr16_-_12809873 | 0.65 |
ENSDART00000146997
ENSDART00000178291 ENSDART00000007842 |
isoc2
|
isochorismatase domain containing 2 |
| chr16_-_36834505 | 0.64 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr21_+_20771082 | 0.64 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr13_-_37642890 | 0.64 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr16_-_41503925 | 0.64 |
ENSDART00000029492
|
cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr22_+_36914636 | 0.63 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr21_-_30545121 | 0.63 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr23_-_39636195 | 0.62 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr19_+_712127 | 0.61 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr18_-_16801033 | 0.60 |
ENSDART00000100100
|
admb
|
adrenomedullin b |
| chr3_-_44059902 | 0.60 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr5_-_25582721 | 0.60 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr18_+_10820430 | 0.60 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr5_-_25583125 | 0.60 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr9_+_41218967 | 0.59 |
ENSDART00000000280
ENSDART00000145674 |
stat1b
|
signal transducer and activator of transcription 1b |
| chr13_+_6092135 | 0.59 |
ENSDART00000162738
|
fam120b
|
family with sequence similarity 120B |
| chr6_-_8498908 | 0.59 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr5_+_13359146 | 0.58 |
ENSDART00000179839
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr19_+_43359075 | 0.58 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr4_-_18436899 | 0.57 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr24_+_13635108 | 0.57 |
ENSDART00000183008
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr1_+_10129099 | 0.57 |
ENSDART00000187740
|
rbm46
|
RNA binding motif protein 46 |
| chr12_+_19036380 | 0.56 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr8_+_19548985 | 0.56 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr1_-_2334254 | 0.56 |
ENSDART00000144500
|
si:ch211-235f1.3
|
si:ch211-235f1.3 |
| chr8_-_43120844 | 0.55 |
ENSDART00000193242
|
ccdc92
|
coiled-coil domain containing 92 |
| chr18_+_3332999 | 0.55 |
ENSDART00000160857
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr18_-_14860435 | 0.55 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr21_-_20932603 | 0.53 |
ENSDART00000138155
ENSDART00000079709 |
c6
|
complement component 6 |
| chr16_+_54513267 | 0.52 |
ENSDART00000161923
|
CABZ01088508.1
|
|
| chr9_-_6372535 | 0.52 |
ENSDART00000149189
|
ecrg4a
|
esophageal cancer related gene 4a |
| chr18_+_30567945 | 0.51 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr25_-_4530085 | 0.51 |
ENSDART00000144907
|
pidd1
|
p53-induced death domain protein 1 |
| chr4_+_76735113 | 0.51 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr24_-_21674950 | 0.51 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr10_-_11012000 | 0.51 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr9_-_7652792 | 0.50 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr10_-_27223827 | 0.50 |
ENSDART00000185138
|
auts2a
|
autism susceptibility candidate 2a |
| chr22_+_20427170 | 0.50 |
ENSDART00000136744
|
foxq2
|
forkhead box Q2 |
| chr6_-_35106425 | 0.50 |
ENSDART00000165139
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr15_-_32383340 | 0.49 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr14_-_36799280 | 0.49 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr24_+_9590188 | 0.49 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr1_-_53504603 | 0.49 |
ENSDART00000162025
ENSDART00000100788 |
commd1
|
copper metabolism (Murr1) domain containing 1 |
| chr15_+_6661343 | 0.48 |
ENSDART00000160136
|
nop53
|
NOP53 ribosome biogenesis factor |
| chr13_-_37631092 | 0.48 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr20_-_23440955 | 0.47 |
ENSDART00000153386
|
slc10a4
|
solute carrier family 10, member 4 |
| chr12_+_7491690 | 0.47 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr4_-_779796 | 0.47 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr23_-_40792128 | 0.46 |
ENSDART00000145360
|
si:dkey-194e6.2
|
si:dkey-194e6.2 |
| chr5_-_42883761 | 0.46 |
ENSDART00000167374
|
BX323596.2
|
|
| chr17_+_10318071 | 0.46 |
ENSDART00000161844
|
foxa1
|
forkhead box A1 |
| chr9_-_23922011 | 0.45 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr9_-_32813177 | 0.45 |
ENSDART00000012694
|
mxc
|
myxovirus (influenza virus) resistance C |
| chr2_+_22659787 | 0.45 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr24_+_13635387 | 0.45 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr1_-_9195629 | 0.45 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr7_-_28696556 | 0.45 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr6_-_12456077 | 0.44 |
ENSDART00000190107
|
gpd2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr25_+_6186823 | 0.44 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr19_+_7759354 | 0.44 |
ENSDART00000151400
|
ubap2l
|
ubiquitin associated protein 2-like |
| chr3_-_7524363 | 0.42 |
ENSDART00000162970
|
znf1001
|
zinc finger protein 1001 |
| chr1_-_10071422 | 0.41 |
ENSDART00000135522
ENSDART00000033118 |
fga
|
fibrinogen alpha chain |
| chr5_+_32009956 | 0.41 |
ENSDART00000188482
|
scai
|
suppressor of cancer cell invasion |
| chr20_-_38446891 | 0.41 |
ENSDART00000192013
|
itpkb
|
inositol-trisphosphate 3-kinase B |
| chr14_+_6159162 | 0.41 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr13_-_49819027 | 0.40 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr15_+_23550890 | 0.39 |
ENSDART00000009796
ENSDART00000152720 |
MARK4
|
si:dkey-31m14.7 |
| chr4_+_359970 | 0.38 |
ENSDART00000139832
|
tmem181
|
transmembrane protein 181 |
| chr21_-_22715297 | 0.37 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr23_-_40776046 | 0.36 |
ENSDART00000136230
|
DUPD1 (1 of many)
|
si:dkeyp-27c8.1 |
| chr16_+_30575901 | 0.36 |
ENSDART00000077231
|
mc2r
|
melanocortin 2 receptor |
| chr15_-_35246742 | 0.36 |
ENSDART00000131479
|
mff
|
mitochondrial fission factor |
| chr15_-_30832897 | 0.36 |
ENSDART00000152330
|
msi2b
|
musashi RNA-binding protein 2b |
| chr7_+_52766211 | 0.35 |
ENSDART00000186191
|
ppip5k1a
|
diphosphoinositol pentakisphosphate kinase 1a |
| chr24_-_22702017 | 0.35 |
ENSDART00000179403
|
ctnnd2a
|
catenin (cadherin-associated protein), delta 2a |
| chr15_-_31067589 | 0.34 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr9_-_54304684 | 0.34 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr14_+_11457500 | 0.33 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr1_-_49225890 | 0.33 |
ENSDART00000111598
|
cxcl18b
|
chemokine (C-X-C motif) ligand 18b |
| chr9_+_29585943 | 0.33 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr16_-_29458806 | 0.32 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr15_-_3282220 | 0.32 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr11_-_22303678 | 0.32 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr4_-_9810999 | 0.32 |
ENSDART00000146858
|
si:dkeyp-27e10.3
|
si:dkeyp-27e10.3 |
| chr6_+_18321627 | 0.31 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr12_-_3252263 | 0.31 |
ENSDART00000170632
|
gngt2b
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2b |
| chr16_-_36099492 | 0.31 |
ENSDART00000180905
|
CU499336.2
|
|
| chr3_+_24511959 | 0.30 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr20_+_53521648 | 0.30 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr19_-_26235219 | 0.30 |
ENSDART00000104008
|
dtnbp1b
|
dystrobrevin binding protein 1b |
| chr16_-_21668082 | 0.30 |
ENSDART00000088513
|
gnl1
|
guanine nucleotide binding protein-like 1 |
| chrM_+_4993 | 0.29 |
ENSDART00000093600
|
mt-nd2
|
NADH dehydrogenase 2, mitochondrial |
| chr23_-_19682971 | 0.29 |
ENSDART00000048891
|
zgc:193598
|
zgc:193598 |
| chr2_-_10098191 | 0.29 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr17_-_7861219 | 0.29 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr1_-_7582859 | 0.28 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr17_+_6563307 | 0.28 |
ENSDART00000156454
|
adgrf3a
|
adhesion G protein-coupled receptor F3a |
| chr1_-_54997746 | 0.28 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr7_+_41313568 | 0.28 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr14_+_6159356 | 0.28 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr17_-_31344400 | 0.28 |
ENSDART00000154013
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr2_+_4402765 | 0.27 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr16_-_45398408 | 0.27 |
ENSDART00000004052
|
rgl2
|
ral guanine nucleotide dissociation stimulator-like 2 |
| chr23_+_7379728 | 0.27 |
ENSDART00000012194
|
gata5
|
GATA binding protein 5 |
| chr19_-_35596207 | 0.27 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr21_+_1568044 | 0.26 |
ENSDART00000193570
ENSDART00000151601 |
rab27b
|
RAB27B, member RAS oncogene family |
| chr11_+_36777613 | 0.26 |
ENSDART00000065644
ENSDART00000158151 |
parapinopsinb
|
parapinopsin b |
| chr2_-_25140022 | 0.26 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr9_+_6934491 | 0.25 |
ENSDART00000114323
|
mfsd9
|
major facilitator superfamily domain containing 9 |
| chr1_-_46343999 | 0.25 |
ENSDART00000145117
ENSDART00000193233 |
atp11a
|
ATPase phospholipid transporting 11A |
| chr2_+_38261748 | 0.25 |
ENSDART00000076478
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr7_+_48319916 | 0.25 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr20_-_25412181 | 0.25 |
ENSDART00000062780
|
itsn2a
|
intersectin 2a |
| chr7_-_5191797 | 0.25 |
ENSDART00000172955
|
si:ch73-223f5.1
|
si:ch73-223f5.1 |
| chr20_-_47188966 | 0.25 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr2_+_4207209 | 0.24 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr5_+_25072894 | 0.24 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr17_-_26604946 | 0.24 |
ENSDART00000087062
|
fam149b1
|
family with sequence similarity 149, member B1 |
| chr16_-_1502699 | 0.24 |
ENSDART00000187189
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr10_+_22134606 | 0.24 |
ENSDART00000155228
|
si:dkey-4c2.11
|
si:dkey-4c2.11 |
| chr4_+_76575585 | 0.24 |
ENSDART00000131588
|
ms4a17a.11
|
membrane-spanning 4-domains, subfamily A, member 17A.11 |
| chr6_+_11989537 | 0.24 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr9_-_23922778 | 0.24 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr21_-_20840714 | 0.23 |
ENSDART00000144861
ENSDART00000139430 |
c6
|
complement component 6 |
| chr1_+_30083578 | 0.23 |
ENSDART00000101779
ENSDART00000143639 |
vps8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr6_-_8498676 | 0.23 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr20_-_39273987 | 0.23 |
ENSDART00000127173
|
clu
|
clusterin |
| chr20_+_25552057 | 0.23 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr5_+_1911814 | 0.23 |
ENSDART00000172233
|
si:ch73-55i23.1
|
si:ch73-55i23.1 |
| chr7_+_41314862 | 0.23 |
ENSDART00000185198
|
zgc:165532
|
zgc:165532 |
| chr14_-_38828057 | 0.23 |
ENSDART00000186088
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr6_-_40286633 | 0.22 |
ENSDART00000033844
|
col7a1
|
collagen, type VII, alpha 1 |
| chr22_-_834106 | 0.22 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr19_+_10587307 | 0.22 |
ENSDART00000131225
|
si:dkey-211g8.4
|
si:dkey-211g8.4 |
| chr11_+_25328199 | 0.22 |
ENSDART00000141478
ENSDART00000112209 |
fam83d
|
family with sequence similarity 83, member D |
| chr7_-_28658143 | 0.22 |
ENSDART00000173556
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr22_+_26263290 | 0.21 |
ENSDART00000184840
|
BX649315.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of stat1a+stat2-1_stat1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.4 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.3 | 1.0 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.3 | 1.0 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.3 | 1.2 | GO:0060467 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.3 | 1.2 | GO:0015871 | choline transport(GO:0015871) |
| 0.3 | 0.9 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.3 | 0.8 | GO:0009595 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 0.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 1.2 | GO:0045624 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 | 0.5 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.2 | 0.5 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.2 | 0.5 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.6 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 1.0 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.6 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.1 | 0.7 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.3 | GO:1990575 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.1 | 1.2 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.3 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.1 | 0.4 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.1 | 0.5 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 1.0 | GO:0051121 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.9 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.1 | 0.8 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 1.4 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.1 | 0.3 | GO:0033363 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.1 | 0.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.3 | GO:0035739 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.3 | GO:0048618 | post-embryonic foregut morphogenesis(GO:0048618) |
| 0.1 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.3 | GO:0016038 | absorption of visible light(GO:0016038) |
| 0.1 | 3.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 0.4 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.1 | 0.1 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.6 | GO:0034340 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.1 | 0.3 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.2 | GO:0060923 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.1 | 0.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 3.1 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.5 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.7 | GO:0002548 | monocyte chemotaxis(GO:0002548) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.6 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.7 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0019628 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.0 | 0.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.7 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.4 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.0 | 0.1 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.0 | 0.8 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.5 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 1.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.2 | GO:1902808 | positive regulation of cell cycle G1/S phase transition(GO:1902808) |
| 0.0 | 0.4 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.1 | GO:0002526 | acute inflammatory response(GO:0002526) acute-phase response(GO:0006953) |
| 0.0 | 0.2 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.0 | 0.3 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 1.4 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.1 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.6 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0002551 | mast cell chemotaxis(GO:0002551) positive regulation of leukocyte chemotaxis(GO:0002690) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0002834 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.9 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.3 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 0.2 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.4 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 1.2 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 2.1 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 1.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.4 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0060975 | cardioblast migration(GO:0003260) cardioblast migration to the midline involved in heart field formation(GO:0060975) |
| 0.0 | 0.3 | GO:0050795 | regulation of behavior(GO:0050795) |
| 0.0 | 0.1 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0071219 | cellular response to molecule of bacterial origin(GO:0071219) cellular response to lipopolysaccharide(GO:0071222) |
| 0.0 | 4.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0015746 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.4 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.4 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 1.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 3.5 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.7 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.8 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.8 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0030286 | dynein complex(GO:0030286) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.3 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.4 | 1.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.3 | 1.2 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.3 | 0.8 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 1.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 2.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.6 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.1 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.4 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 1.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.5 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.4 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 0.5 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.9 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.6 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.8 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.4 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0050254 | rhodopsin kinase activity(GO:0050254) |
| 0.0 | 1.1 | GO:0015278 | calcium-release channel activity(GO:0015278) |
| 0.0 | 0.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 1.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.3 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0031769 | glucagon receptor binding(GO:0031769) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 1.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.0 | 0.7 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.3 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.6 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.2 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.7 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 1.3 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |