Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
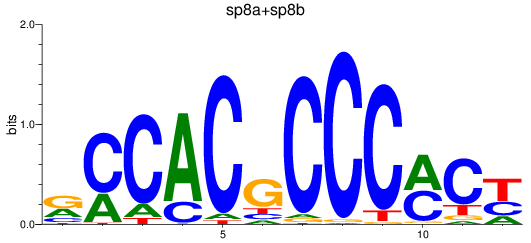
Results for sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
Z-value: 1.39
Transcription factors associated with sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp8a
|
ENSDARG00000011870 | sp8 transcription factor a |
|
sp8b
|
ENSDARG00000056666 | sp8 transcription factor b |
|
sp4
|
ENSDARG00000005186 | sp4 transcription factor |
|
sp3a
|
ENSDARG00000001549 | sp3a transcription factor |
|
rsl1d1
|
ENSDARG00000005846 | Sp5 transcription factor b |
|
klf4
|
ENSDARG00000079922 | Kruppel-like factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp8a | dr11_v1_chr19_-_2317558_2317558 | 0.88 | 1.2e-06 | Click! |
| sp3a | dr11_v1_chr9_+_2762270_2762415 | 0.83 | 2.0e-05 | Click! |
| klf4 | dr11_v1_chr21_-_435466_435466 | -0.76 | 2.3e-04 | Click! |
| sp4 | dr11_v1_chr19_+_2546775_2546775 | 0.75 | 3.4e-04 | Click! |
| FO704755.1 | dr11_v1_chr6_+_3809346_3809346 | -0.38 | 1.2e-01 | Click! |
| sp8b | dr11_v1_chr16_+_19536614_19536761 | -0.27 | 2.9e-01 | Click! |
Activity profile of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4 motif
Sorted Z-values of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_410728 | 7.20 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr7_+_1473929 | 6.02 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr2_+_25658112 | 5.69 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr22_+_835728 | 5.31 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr10_-_34002185 | 5.20 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr23_-_31645760 | 5.11 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr16_-_7793457 | 5.09 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr2_+_25657958 | 4.97 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr19_+_26340736 | 4.94 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr20_-_182841 | 4.94 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr18_-_127558 | 4.88 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr19_-_11031145 | 4.86 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr7_+_38349667 | 4.79 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr3_-_40054615 | 4.77 |
ENSDART00000003511
ENSDART00000102540 ENSDART00000146121 |
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr12_+_46634736 | 4.55 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr8_-_410199 | 4.17 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr18_-_127873 | 4.09 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr17_-_18898115 | 3.97 |
ENSDART00000028044
|
galcb
|
galactosylceramidase b |
| chr5_-_13086616 | 3.80 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr11_-_34577034 | 3.75 |
ENSDART00000133302
ENSDART00000184367 |
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr2_+_68789 | 3.70 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr21_-_32060993 | 3.68 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr16_+_53455638 | 3.62 |
ENSDART00000045792
ENSDART00000154189 |
rbm24b
|
RNA binding motif protein 24b |
| chr10_-_41352502 | 3.62 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr17_+_25856671 | 3.60 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr3_-_50998577 | 3.45 |
ENSDART00000157735
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr8_+_47219107 | 3.44 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr12_-_37449396 | 3.43 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr3_+_32410746 | 3.42 |
ENSDART00000025496
|
rras
|
RAS related |
| chr7_+_40083601 | 3.30 |
ENSDART00000099046
|
zgc:112356
|
zgc:112356 |
| chr10_-_44017642 | 3.29 |
ENSDART00000135240
ENSDART00000014669 |
acads
|
acyl-CoA dehydrogenase short chain |
| chr21_-_43398122 | 3.28 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr19_-_867071 | 3.20 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr21_-_13856689 | 3.14 |
ENSDART00000102197
|
fam129ba
|
family with sequence similarity 129, member Ba |
| chr7_+_55518519 | 3.14 |
ENSDART00000098476
ENSDART00000149915 |
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr1_-_34450784 | 3.12 |
ENSDART00000140515
|
lmo7b
|
LIM domain 7b |
| chr5_-_69482891 | 3.08 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr5_-_23696926 | 3.08 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr3_-_15470944 | 3.01 |
ENSDART00000185302
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr1_-_34450622 | 2.97 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr6_-_7776612 | 2.92 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr1_+_45707219 | 2.92 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr10_-_2942900 | 2.84 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr20_+_46741074 | 2.83 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr7_-_19332293 | 2.79 |
ENSDART00000169668
ENSDART00000137575 ENSDART00000090406 |
dock11
|
dedicator of cytokinesis 11 |
| chr25_+_3104959 | 2.78 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr19_+_46113828 | 2.77 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr21_-_43398457 | 2.76 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr19_-_6193448 | 2.73 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr22_+_883678 | 2.73 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr6_+_41503854 | 2.72 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr5_+_66132394 | 2.71 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr14_+_7048930 | 2.68 |
ENSDART00000109138
|
hbegfa
|
heparin-binding EGF-like growth factor a |
| chr22_+_11756040 | 2.68 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr16_+_30002605 | 2.66 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr24_+_35387517 | 2.65 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr23_-_452365 | 2.65 |
ENSDART00000146776
|
tspan2b
|
tetraspanin 2b |
| chr23_+_12160900 | 2.59 |
ENSDART00000136046
|
ppp1r3da
|
protein phosphatase 1, regulatory subunit 3Da |
| chr17_+_51746830 | 2.54 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr2_-_11027258 | 2.54 |
ENSDART00000081072
ENSDART00000193824 ENSDART00000187036 ENSDART00000097741 |
ssbp3a
|
single stranded DNA binding protein 3a |
| chr9_-_11263228 | 2.52 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr10_+_39199547 | 2.50 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr2_-_39675829 | 2.49 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr22_-_38459316 | 2.47 |
ENSDART00000149683
ENSDART00000098461 |
ptk7a
|
protein tyrosine kinase 7a |
| chr15_+_46344655 | 2.45 |
ENSDART00000155893
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr2_-_47620806 | 2.44 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr19_-_6193067 | 2.42 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr2_+_35880600 | 2.42 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr23_-_10177442 | 2.39 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr19_-_2029777 | 2.37 |
ENSDART00000128639
|
CABZ01071939.1
|
|
| chr22_+_336256 | 2.36 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr5_+_4533244 | 2.33 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr2_+_35603637 | 2.31 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr20_+_51061695 | 2.29 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr23_+_2825940 | 2.29 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr25_+_6122823 | 2.26 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr20_-_14781904 | 2.26 |
ENSDART00000187200
ENSDART00000179912 ENSDART00000160481 ENSDART00000026969 |
suco
|
SUN domain containing ossification factor |
| chr22_-_20924747 | 2.26 |
ENSDART00000185845
ENSDART00000179672 |
ell
|
elongation factor RNA polymerase II |
| chr22_-_20924564 | 2.26 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr4_-_4612116 | 2.25 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr3_-_32362872 | 2.24 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr8_+_2656231 | 2.21 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr16_+_43077909 | 2.21 |
ENSDART00000014140
|
rundc3b
|
RUN domain containing 3b |
| chr25_+_7492663 | 2.20 |
ENSDART00000166496
|
cat
|
catalase |
| chr3_+_31925067 | 2.20 |
ENSDART00000127330
ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr9_-_34269066 | 2.20 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr23_+_30730121 | 2.16 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr13_+_421231 | 2.15 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr22_-_10539180 | 2.12 |
ENSDART00000131217
|
ippk
|
inositol 1,3,4,5,6-pentakisphosphate 2-kinase |
| chr7_+_67325933 | 2.10 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr4_+_9011825 | 2.07 |
ENSDART00000058007
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr19_+_47290287 | 2.06 |
ENSDART00000078382
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr8_-_4618653 | 2.05 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr13_+_47821524 | 2.05 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr2_-_32513538 | 2.05 |
ENSDART00000056640
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr4_+_9011448 | 2.04 |
ENSDART00000192357
|
samm50l
|
sorting and assembly machinery component 50 homolog, like |
| chr22_-_10752471 | 2.04 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr21_-_14692119 | 2.03 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr6_+_3334392 | 2.03 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr13_+_42309688 | 2.02 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr22_+_24559947 | 2.01 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr9_-_32343673 | 2.00 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr13_-_18691041 | 2.00 |
ENSDART00000057867
|
sfxn3
|
sideroflexin 3 |
| chr9_-_54840124 | 1.99 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr2_-_37478418 | 1.98 |
ENSDART00000146103
|
dapk3
|
death-associated protein kinase 3 |
| chr23_-_10745288 | 1.98 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr19_+_43341115 | 1.97 |
ENSDART00000145846
ENSDART00000102384 |
sesn2
|
sestrin 2 |
| chr23_-_17429775 | 1.97 |
ENSDART00000043076
|
ppdpfb
|
pancreatic progenitor cell differentiation and proliferation factor b |
| chr21_-_39024754 | 1.96 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr8_-_53044300 | 1.94 |
ENSDART00000191653
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr22_-_6562618 | 1.94 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr11_-_12158412 | 1.93 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr15_-_45538773 | 1.91 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr12_+_48340133 | 1.91 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr3_+_22984098 | 1.91 |
ENSDART00000043190
|
lsm12a
|
LSM12 homolog a |
| chr18_-_22094102 | 1.89 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr19_+_43341424 | 1.89 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr19_+_46237665 | 1.88 |
ENSDART00000159391
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr4_-_5018705 | 1.87 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr10_-_244745 | 1.87 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr9_+_30421489 | 1.85 |
ENSDART00000145025
ENSDART00000132058 |
zgc:113314
|
zgc:113314 |
| chr19_+_9111550 | 1.85 |
ENSDART00000088336
|
setdb1a
|
SET domain, bifurcated 1a |
| chr2_+_59015878 | 1.85 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr24_+_39277043 | 1.84 |
ENSDART00000165458
|
MPRIP
|
si:ch73-103b11.2 |
| chr16_-_31351419 | 1.84 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr10_+_6383270 | 1.84 |
ENSDART00000170548
|
zgc:114200
|
zgc:114200 |
| chr22_-_21676364 | 1.83 |
ENSDART00000183668
|
tle2b
|
transducin like enhancer of split 2b |
| chr17_+_17804752 | 1.82 |
ENSDART00000123350
|
sptlc2a
|
serine palmitoyltransferase, long chain base subunit 2a |
| chr15_+_29024895 | 1.82 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr15_+_46853252 | 1.79 |
ENSDART00000186040
|
zgc:153039
|
zgc:153039 |
| chr13_+_49727333 | 1.78 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr4_-_5019113 | 1.77 |
ENSDART00000189321
ENSDART00000081990 |
strip2
|
striatin interacting protein 2 |
| chr16_-_21489514 | 1.77 |
ENSDART00000149525
ENSDART00000148517 ENSDART00000146914 ENSDART00000186493 ENSDART00000193081 ENSDART00000186017 |
mpp6a
|
membrane protein, palmitoylated 6a (MAGUK p55 subfamily member 6) |
| chr18_-_5248365 | 1.77 |
ENSDART00000082506
ENSDART00000082504 ENSDART00000097960 |
myef2
|
myelin expression factor 2 |
| chr10_+_22034477 | 1.76 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr3_-_15131438 | 1.76 |
ENSDART00000131720
|
xpo6
|
exportin 6 |
| chr2_+_1988036 | 1.75 |
ENSDART00000155956
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr8_-_32385989 | 1.74 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr10_-_35257458 | 1.74 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr24_-_36238054 | 1.72 |
ENSDART00000155725
|
tmem241
|
transmembrane protein 241 |
| chr20_-_23254876 | 1.71 |
ENSDART00000141510
|
ociad1
|
OCIA domain containing 1 |
| chr23_+_35426404 | 1.70 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr7_-_64971839 | 1.68 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr14_-_24277805 | 1.68 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr21_+_21279159 | 1.67 |
ENSDART00000148346
|
itpkca
|
inositol-trisphosphate 3-kinase Ca |
| chr9_+_4378153 | 1.66 |
ENSDART00000191264
ENSDART00000182384 |
kalrna
|
kalirin RhoGEF kinase a |
| chr7_-_4125021 | 1.66 |
ENSDART00000167182
ENSDART00000173696 |
zgc:55733
|
zgc:55733 |
| chr22_+_30184039 | 1.66 |
ENSDART00000049075
|
add3a
|
adducin 3 (gamma) a |
| chr2_-_37134169 | 1.65 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr15_-_25094026 | 1.65 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr8_+_23165749 | 1.63 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr9_-_264173 | 1.63 |
ENSDART00000166231
ENSDART00000165585 |
pcbp2
|
poly(rC) binding protein 2 |
| chr10_-_32851847 | 1.61 |
ENSDART00000134255
|
trim37
|
tripartite motif containing 37 |
| chr4_+_2637947 | 1.61 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr7_+_71764883 | 1.61 |
ENSDART00000166865
|
myl12.1
|
myosin, light chain 12, genome duplicate 1 |
| chr15_+_29025090 | 1.61 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr24_-_28333029 | 1.60 |
ENSDART00000149015
ENSDART00000129174 |
prkag2a
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit a |
| chr18_+_3634652 | 1.60 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr7_-_30492261 | 1.60 |
ENSDART00000173954
|
adam10a
|
ADAM metallopeptidase domain 10a |
| chr6_+_32326074 | 1.60 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr2_-_21438492 | 1.60 |
ENSDART00000046098
|
plcd1b
|
phospholipase C, delta 1b |
| chr19_+_30867845 | 1.60 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr16_-_8120203 | 1.59 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr20_-_2361226 | 1.57 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr5_+_43458304 | 1.57 |
ENSDART00000051114
|
rhobtb2a
|
Rho-related BTB domain containing 2a |
| chr15_-_23942861 | 1.57 |
ENSDART00000002824
|
appbp2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr3_+_18437397 | 1.56 |
ENSDART00000136243
ENSDART00000184882 ENSDART00000135470 |
tbc1d16
|
TBC1 domain family, member 16 |
| chr3_+_24595922 | 1.56 |
ENSDART00000169405
|
si:dkey-68o6.5
|
si:dkey-68o6.5 |
| chr3_-_6417328 | 1.55 |
ENSDART00000160979
|
jpt1b
|
Jupiter microtubule associated homolog 1b |
| chr12_-_3053873 | 1.54 |
ENSDART00000023796
ENSDART00000137148 |
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr14_+_22132388 | 1.54 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr21_+_6197223 | 1.53 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr1_+_604127 | 1.53 |
ENSDART00000133165
|
jam2a
|
junctional adhesion molecule 2a |
| chr14_+_30291611 | 1.53 |
ENSDART00000173004
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr1_+_44196236 | 1.52 |
ENSDART00000179560
ENSDART00000166324 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr9_+_55536005 | 1.52 |
ENSDART00000192364
|
mxra5b
|
matrix-remodelling associated 5b |
| chr1_+_24469313 | 1.51 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr13_-_17860307 | 1.50 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr3_+_51684963 | 1.50 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr14_-_237130 | 1.50 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr18_+_46151505 | 1.50 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr19_-_18135724 | 1.50 |
ENSDART00000186609
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr7_-_59159253 | 1.49 |
ENSDART00000159285
|
haus6
|
HAUS augmin-like complex, subunit 6 |
| chr21_+_31253048 | 1.48 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr20_+_4157815 | 1.48 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr23_-_37085547 | 1.48 |
ENSDART00000144333
|
ints11
|
integrator complex subunit 11 |
| chr9_+_50110763 | 1.48 |
ENSDART00000162990
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr19_-_31686252 | 1.47 |
ENSDART00000131721
|
ripor2
|
RHO family interacting cell polarization regulator 2 |
| chr6_-_17849786 | 1.46 |
ENSDART00000172709
|
rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr25_+_21098675 | 1.46 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr8_-_2616326 | 1.46 |
ENSDART00000027214
|
slc25a25a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25a |
| chr15_-_19128705 | 1.46 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr7_-_58098814 | 1.46 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr15_-_25093680 | 1.45 |
ENSDART00000062695
|
exo5
|
exonuclease 5 |
| chr7_+_71764665 | 1.45 |
ENSDART00000171996
|
myl12.1
|
myosin, light chain 12, genome duplicate 1 |
| chr12_-_4475890 | 1.44 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr20_-_52928541 | 1.44 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr7_-_30492018 | 1.44 |
ENSDART00000099639
ENSDART00000162705 ENSDART00000173663 |
adam10a
|
ADAM metallopeptidase domain 10a |
| chr7_-_38714544 | 1.43 |
ENSDART00000139382
|
fbxo3
|
F-box protein 3 |
| chr3_-_52899394 | 1.42 |
ENSDART00000128223
|
CHCHD5
|
coiled-coil-helix-coiled-coil-helix domain containing 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp8a+sp8b_sp4_rsl1d1+sp3a_klf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 1.6 | 6.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 1.2 | 3.6 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 1.2 | 3.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.1 | 3.2 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 1.0 | 5.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.9 | 2.7 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.8 | 3.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.8 | 2.3 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.8 | 4.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.7 | 2.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.7 | 6.9 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.7 | 2.0 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.7 | 2.0 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.6 | 0.6 | GO:1905067 | negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.6 | 2.9 | GO:0072104 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.6 | 2.3 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.6 | 2.2 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.5 | 2.7 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.5 | 3.8 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.5 | 7.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.5 | 2.1 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.5 | 2.0 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.5 | 2.0 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.5 | 4.0 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.5 | 1.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.5 | 1.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.5 | 1.4 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.5 | 2.3 | GO:0051148 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.4 | 3.5 | GO:0071295 | cellular response to nutrient(GO:0031670) cellular response to vitamin(GO:0071295) |
| 0.4 | 1.8 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.4 | 0.9 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.4 | 1.3 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.4 | 2.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.4 | 1.6 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.4 | 1.5 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.4 | 5.4 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.4 | 1.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.3 | 1.4 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.3 | 1.4 | GO:2000303 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.3 | 5.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 1.0 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.3 | 1.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.3 | 2.5 | GO:2000095 | cerebrospinal fluid circulation(GO:0090660) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.3 | 3.6 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.3 | 2.7 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.3 | 1.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.3 | 1.1 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.3 | 1.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.3 | 0.8 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.3 | 1.4 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.3 | 3.3 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.3 | 1.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.3 | 3.7 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 0.8 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 1.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.3 | 3.0 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.2 | 1.5 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 2.9 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.2 | 2.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 3.8 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 1.9 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.2 | 2.3 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 1.1 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.2 | 1.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 3.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 0.2 | 1.9 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.0 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.2 | 0.8 | GO:0060300 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.2 | 1.0 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.2 | 1.8 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 1.2 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.2 | 0.6 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.2 | 1.0 | GO:0046416 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.2 | 0.4 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.2 | 4.2 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.2 | 1.5 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 1.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 2.2 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 | 1.8 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 1.4 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.2 | 0.5 | GO:1904869 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.2 | 0.9 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.2 | 0.5 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.2 | 0.7 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 | 0.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.2 | 2.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 2.3 | GO:0038202 | TORC1 signaling(GO:0038202) |
| 0.2 | 1.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.2 | 0.9 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.2 | 0.6 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.2 | 1.3 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.2 | 0.3 | GO:2000391 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.2 | 1.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 0.5 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.2 | 2.4 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.1 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 5.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 1.6 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.6 | GO:0045429 | regulation of nitric oxide biosynthetic process(GO:0045428) positive regulation of nitric oxide biosynthetic process(GO:0045429) positive regulation of reactive oxygen species biosynthetic process(GO:1903428) positive regulation of nitric oxide metabolic process(GO:1904407) |
| 0.1 | 2.2 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 3.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 | 1.0 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.4 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 2.1 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 0.4 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.1 | 6.2 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.5 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.1 | 1.0 | GO:1902623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.5 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.1 | 0.4 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.1 | 0.8 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.1 | 0.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.9 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.1 | 2.5 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.4 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.1 | 1.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.8 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) protein urmylation(GO:0032447) |
| 0.1 | 0.9 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 1.2 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.1 | 1.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.5 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.1 | 3.2 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.1 | 1.8 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 2.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 1.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.7 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.1 | 0.7 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 2.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.1 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.1 | 0.5 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.4 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 0.1 | 3.1 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 11.4 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.1 | 0.8 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.1 | 0.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.1 | 0.1 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.1 | 0.3 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.1 | 1.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 1.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 1.6 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 1.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 3.0 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 0.4 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.4 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.1 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 0.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 1.9 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.7 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.5 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.1 | 1.4 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.4 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.3 | GO:0046104 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.9 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.3 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.1 | 0.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.5 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.3 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 | 0.7 | GO:1903578 | regulation of oxidative phosphorylation(GO:0002082) regulation of nucleoside metabolic process(GO:0009118) regulation of ATP metabolic process(GO:1903578) |
| 0.1 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.6 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.9 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.2 | GO:0031335 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 1.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.1 | 1.2 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 1.3 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.4 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.1 | 1.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.3 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.1 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 1.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 0.2 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.1 | 0.6 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 1.4 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.1 | 3.7 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.1 | 0.2 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.2 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.1 | 0.4 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.0 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.1 | 0.4 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.9 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.1 | 1.3 | GO:0043574 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 1.5 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.9 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.1 | 0.8 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.5 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 1.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 4.8 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.1 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.3 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.2 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 1.4 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.5 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 1.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.5 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 2.2 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.2 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 2.7 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.0 | 2.4 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 2.5 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.2 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.3 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.1 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.3 | GO:0015961 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 1.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.7 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.4 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.2 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.9 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 2.0 | GO:0018198 | peptidyl-cysteine modification(GO:0018198) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.4 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.0 | 0.5 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.2 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 1.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 1.2 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.0 | 2.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 3.2 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.5 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.0 | 0.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.5 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 1.7 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.2 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.3 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 1.9 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 1.0 | GO:0006505 | GPI anchor metabolic process(GO:0006505) |
| 0.0 | 0.2 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.5 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 4.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.4 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.9 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 7.9 | GO:0006644 | phospholipid metabolic process(GO:0006644) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 1.0 | GO:0000077 | DNA damage checkpoint(GO:0000077) |
| 0.0 | 1.0 | GO:0048884 | neuromast development(GO:0048884) |
| 0.0 | 0.7 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.3 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 2.6 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.1 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 2.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.1 | GO:0006210 | thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.0 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.9 | GO:0044042 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0000467 | exonucleolytic trimming involved in rRNA processing(GO:0000459) exonucleolytic trimming to generate mature 3'-end of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000467) |
| 0.0 | 0.2 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.1 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.2 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 3.4 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.2 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.0 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0001736 | establishment of planar polarity(GO:0001736) establishment of tissue polarity(GO:0007164) |
| 0.0 | 0.7 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 0.3 | GO:0038034 | signal transduction in absence of ligand(GO:0038034) negative regulation of mitotic nuclear division(GO:0045839) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.1 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.4 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0006555 | methionine metabolic process(GO:0006555) |
| 0.0 | 0.1 | GO:0045176 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.0 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.1 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 2.0 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.1 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.0 | 0.0 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.6 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.2 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 1.4 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 1.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0003242 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0010573 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 1.2 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.4 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 2.2 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) DNA strand elongation(GO:0022616) |
| 0.0 | 0.1 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.8 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.0 | 0.2 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0098536 | deuterosome(GO:0098536) |
| 0.6 | 4.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.5 | 0.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.5 | 2.0 | GO:1990498 | mitotic spindle microtubule(GO:1990498) |
| 0.5 | 3.6 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.3 | 1.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.3 | 2.0 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 2.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 1.3 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.3 | 2.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.3 | 0.9 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.2 | 1.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 1.5 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 1.4 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.2 | 2.9 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.2 | 0.8 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.2 | 1.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 2.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 1.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.2 | 3.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 3.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 2.4 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.2 | 2.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.8 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 0.6 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.2 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.0 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.6 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 6.1 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.7 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.4 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 1.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 0.6 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 1.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.7 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 3.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.3 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 1.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.6 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 1.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.7 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.1 | 0.3 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 4.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 10.5 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 4.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 6.9 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.8 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.4 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 3.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 3.2 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.1 | 0.5 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 1.2 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 1.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.4 | GO:0016528 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.2 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 3.3 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.0 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.5 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.2 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 4.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 9.1 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 5.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.5 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.8 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.4 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 2.4 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 3.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.7 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0032806 | holo TFIIH complex(GO:0005675) carboxy-terminal domain protein kinase complex(GO:0032806) |
| 0.0 | 0.5 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.9 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.7 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 1.3 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 0.7 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.4 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 10.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 4.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 5.4 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.8 | GO:0005819 | spindle(GO:0005819) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.9 | 2.8 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.9 | 2.8 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.8 | 3.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.8 | 3.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.7 | 2.2 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.7 | 2.2 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.5 | 7.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.5 | 3.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 5.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.5 | 1.4 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.4 | 2.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.4 | 2.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 1.5 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.4 | 2.6 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.4 | 1.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.4 | 2.9 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 3.8 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.3 | 2.0 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.3 | 5.4 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.3 | 1.1 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.3 | 0.9 | GO:0034618 | arginine binding(GO:0034618) |
| 0.3 | 0.8 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.3 | 1.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.3 | 2.5 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.3 | 0.8 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.3 | 0.8 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 2.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.3 | 3.8 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 1.6 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 2.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 2.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.2 | 1.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.2 | 1.5 | GO:0001091 | RNA polymerase II basal transcription factor binding(GO:0001091) |
| 0.2 | 1.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.2 | 2.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 3.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.2 | 2.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.2 | 1.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 2.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.2 | 1.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.2 | 1.0 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.2 | 1.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.2 | 3.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 1.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.0 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.2 | 1.1 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.2 | 0.7 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 2.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 1.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.2 | 4.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 0.6 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.2 | 1.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.2 | 4.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 5.9 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.4 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.4 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.1 | 0.4 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 1.0 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.4 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.1 | 1.0 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.1 | 1.0 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.4 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 3.1 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 1.4 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 1.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.4 | GO:0003868 | 4-hydroxyphenylpyruvate dioxygenase activity(GO:0003868) |
| 0.1 | 1.6 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 1.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.7 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.1 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 2.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 0.3 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.9 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 3.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 2.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 3.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 2.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 3.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 0.3 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.1 | 1.7 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 1.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.8 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.3 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 1.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.8 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.4 | GO:0050333 | thiamin-triphosphatase activity(GO:0050333) |
| 0.1 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.1 | 3.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 1.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.3 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 1.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.5 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.4 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.5 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 6.7 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 3.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.6 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 2.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.5 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.9 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.2 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 0.3 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.2 | GO:0003999 | adenine phosphoribosyltransferase activity(GO:0003999) |
| 0.1 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 1.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.8 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 1.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 1.0 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 1.8 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 1.4 | GO:0035257 | nuclear hormone receptor binding(GO:0035257) |
| 0.0 | 0.2 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.0 | 0.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.5 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.4 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 6.5 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.4 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.9 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.8 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.0 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.4 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 1.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.2 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 11.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 1.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.4 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.0 | 0.8 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.3 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.0 | 1.4 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.0 | 1.9 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.2 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.5 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 5.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 2.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.0 | 0.5 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.6 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.8 | GO:0019212 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 2.5 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 4.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.5 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.4 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.0 | 0.2 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 1.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 0.3 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.1 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 0.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 6.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 1.2 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 14.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.6 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 10.7 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.6 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.3 | 3.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.3 | 4.7 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 2.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 9.1 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 2.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.9 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 0.8 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.6 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 1.9 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 9.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 1.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.2 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.4 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.1 | 2.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.7 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.1 | 2.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.4 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.1 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 1.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 5.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.6 | 5.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.6 | 2.3 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.4 | 2.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.4 | 3.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 4.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.3 | 5.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 7.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.3 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.2 | 3.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 1.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.2 | 4.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 1.0 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.2 | 2.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 3.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 1.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 1.6 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 3.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 2.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.8 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 3.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 2.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.1 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 1.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.9 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 1.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 4.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 0.9 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.5 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.1 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.1 | 1.3 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.1 | 1.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 2.6 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 1.1 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.9 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.8 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 2.5 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 3.5 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.1 | REACTOME SEMAPHORIN INTERACTIONS | Genes involved in Semaphorin interactions |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 3.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.7 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |