Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
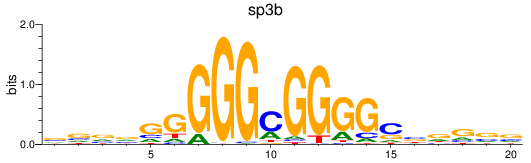
Results for sp3b
Z-value: 0.83
Transcription factors associated with sp3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp3b
|
ENSDARG00000007812 | Sp3b transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp3b | dr11_v1_chr6_-_10728057_10728057 | -0.89 | 8.2e-07 | Click! |
Activity profile of sp3b motif
Sorted Z-values of sp3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_44924289 | 4.57 |
ENSDART00000171267
|
tuba7l
|
tubulin, alpha 7 like |
| chr10_-_641609 | 4.13 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr6_-_60104628 | 4.03 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr20_-_147574 | 3.35 |
ENSDART00000104762
ENSDART00000131635 |
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr2_-_44183451 | 3.32 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr7_-_16598212 | 3.10 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr2_-_44183613 | 2.85 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr6_+_475264 | 2.68 |
ENSDART00000193615
|
LO017974.1
|
|
| chr3_-_16227683 | 2.61 |
ENSDART00000111707
|
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr15_+_19838458 | 2.27 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr16_-_45058919 | 2.15 |
ENSDART00000177134
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr6_+_7249531 | 2.05 |
ENSDART00000125912
ENSDART00000083424 ENSDART00000049695 ENSDART00000136088 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr24_+_33622769 | 2.03 |
ENSDART00000079342
|
dnah5l
|
dynein, axonemal, heavy chain 5 like |
| chr19_-_31802296 | 2.01 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr14_-_2933185 | 1.98 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr3_+_1211242 | 1.93 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr6_+_29217392 | 1.91 |
ENSDART00000006386
|
atp1b1a
|
ATPase Na+/K+ transporting subunit beta 1a |
| chr7_-_10560964 | 1.88 |
ENSDART00000172761
ENSDART00000170476 |
mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr5_-_68022631 | 1.86 |
ENSDART00000143199
|
wasf3a
|
WAS protein family, member 3a |
| chr7_+_34688527 | 1.86 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr19_+_935565 | 1.81 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr19_+_48111285 | 1.77 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr25_+_1732838 | 1.76 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr8_+_27555314 | 1.75 |
ENSDART00000135568
ENSDART00000016696 |
rhocb
|
ras homolog family member Cb |
| chr20_-_7080427 | 1.73 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr21_-_43015383 | 1.73 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr1_-_59232267 | 1.72 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr23_+_17220986 | 1.71 |
ENSDART00000054761
|
nol4lb
|
nucleolar protein 4-like b |
| chr19_+_636886 | 1.67 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr20_-_53981626 | 1.66 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr10_-_39154778 | 1.64 |
ENSDART00000186811
|
slc37a4b
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4b |
| chr1_-_58562129 | 1.64 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr13_-_36703164 | 1.63 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr17_-_4395373 | 1.59 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr7_+_568819 | 1.57 |
ENSDART00000173716
|
nrxn2b
|
neurexin 2b |
| chr3_-_18030938 | 1.56 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr6_+_9241121 | 1.54 |
ENSDART00000064989
|
pimr70
|
Pim proto-oncogene, serine/threonine kinase, related 70 |
| chr7_+_529522 | 1.53 |
ENSDART00000190811
|
nrxn2b
|
neurexin 2b |
| chr3_+_12554801 | 1.51 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr3_+_14388010 | 1.50 |
ENSDART00000171726
ENSDART00000165452 |
tmem56b
|
transmembrane protein 56b |
| chr5_-_1047222 | 1.47 |
ENSDART00000181112
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr16_+_4838808 | 1.47 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr19_-_12145765 | 1.45 |
ENSDART00000032474
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr19_+_4912817 | 1.44 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr13_-_33683889 | 1.43 |
ENSDART00000136820
ENSDART00000065435 |
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr3_-_22829710 | 1.42 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr3_-_16289826 | 1.40 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr14_-_9522364 | 1.37 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr19_-_12145390 | 1.32 |
ENSDART00000143087
|
pabpc1b
|
poly A binding protein, cytoplasmic 1 b |
| chr12_+_49125510 | 1.31 |
ENSDART00000185804
|
FO704607.1
|
|
| chr13_+_37022601 | 1.30 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr9_-_24413008 | 1.29 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr1_-_58561963 | 1.29 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr7_-_24236364 | 1.27 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr22_+_2860260 | 1.23 |
ENSDART00000106221
|
si:dkey-20i20.2
|
si:dkey-20i20.2 |
| chr13_-_5978433 | 1.21 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr14_-_24391424 | 1.20 |
ENSDART00000113376
ENSDART00000126894 |
fam13b
|
family with sequence similarity 13, member B |
| chr10_-_27197044 | 1.20 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr25_+_19149241 | 1.20 |
ENSDART00000184982
ENSDART00000067324 |
mfge8b
|
milk fat globule-EGF factor 8 protein b |
| chr16_+_4839078 | 1.19 |
ENSDART00000150111
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr5_-_40910749 | 1.19 |
ENSDART00000083467
ENSDART00000133183 |
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_9893554 | 1.14 |
ENSDART00000064979
|
pimr74
|
Pim proto-oncogene, serine/threonine kinase, related 74 |
| chr4_-_18595525 | 1.13 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr5_-_46273938 | 1.13 |
ENSDART00000080033
|
si:ch211-130m23.3
|
si:ch211-130m23.3 |
| chr24_+_40473032 | 1.09 |
ENSDART00000084238
ENSDART00000178508 |
CABZ01076968.1
|
|
| chr1_-_669717 | 1.09 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr13_+_41022502 | 1.06 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr9_-_9671244 | 1.05 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr5_-_22619879 | 1.02 |
ENSDART00000051623
|
zgc:113208
|
zgc:113208 |
| chr22_+_10752511 | 1.02 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_-_45349849 | 1.01 |
ENSDART00000183036
|
CABZ01068367.1
|
Danio rerio uncharacterized LOC100332446 (LOC100332446), mRNA. |
| chr25_-_8138122 | 1.01 |
ENSDART00000104659
|
sergef
|
secretion regulating guanine nucleotide exchange factor |
| chr4_+_72742212 | 1.00 |
ENSDART00000171021
|
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr17_+_43889371 | 1.00 |
ENSDART00000156871
ENSDART00000154702 |
msh4
|
mutS homolog 4 |
| chr22_+_10752787 | 0.98 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr6_-_43922813 | 0.97 |
ENSDART00000123341
|
prok2
|
prokineticin 2 |
| chr3_-_16227490 | 0.96 |
ENSDART00000057159
ENSDART00000130611 ENSDART00000012835 |
cacnb1
|
calcium channel, voltage-dependent, beta 1 subunit |
| chr2_+_55365727 | 0.96 |
ENSDART00000162943
|
FP245456.1
|
|
| chr1_-_51157660 | 0.96 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr1_+_144284 | 0.95 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr4_-_15603511 | 0.94 |
ENSDART00000122520
ENSDART00000162356 |
chchd3a
|
coiled-coil-helix-coiled-coil-helix domain containing 3a |
| chr3_-_62380146 | 0.94 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr7_-_16597130 | 0.94 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr7_-_12968689 | 0.93 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr17_-_43466317 | 0.92 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr7_-_18168493 | 0.92 |
ENSDART00000127428
|
peli3
|
pellino E3 ubiquitin protein ligase family member 3 |
| chr18_-_15467446 | 0.92 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr18_-_50799510 | 0.91 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr6_-_442163 | 0.89 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr17_+_8925232 | 0.89 |
ENSDART00000036668
|
psmc1a
|
proteasome 26S subunit, ATPase 1a |
| chr4_-_37766174 | 0.88 |
ENSDART00000170066
|
si:dkey-207l24.2
|
si:dkey-207l24.2 |
| chr16_+_11029762 | 0.88 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr23_-_46201008 | 0.87 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr24_-_4450238 | 0.87 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr1_-_18803919 | 0.86 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr10_+_10210455 | 0.85 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr11_-_236766 | 0.85 |
ENSDART00000163978
|
dusp7
|
dual specificity phosphatase 7 |
| chr12_+_33919502 | 0.84 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr22_+_13886821 | 0.84 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr5_+_51443009 | 0.83 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr8_+_23213320 | 0.83 |
ENSDART00000032996
ENSDART00000137536 |
ppdpfa
|
pancreatic progenitor cell differentiation and proliferation factor a |
| chr21_+_28445052 | 0.83 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr15_-_47956388 | 0.82 |
ENSDART00000116506
|
si:ch1073-111c8.3
|
si:ch1073-111c8.3 |
| chr3_-_5964557 | 0.82 |
ENSDART00000184738
|
BX284638.1
|
|
| chr6_+_58915889 | 0.82 |
ENSDART00000083628
|
ddit3
|
DNA-damage-inducible transcript 3 |
| chr7_-_41468942 | 0.82 |
ENSDART00000174087
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr19_+_9186175 | 0.82 |
ENSDART00000039325
|
hcn3
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 3 |
| chr14_+_24283915 | 0.81 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr10_-_28835771 | 0.81 |
ENSDART00000192220
ENSDART00000188436 |
alcama
|
activated leukocyte cell adhesion molecule a |
| chr10_+_33895315 | 0.81 |
ENSDART00000142881
|
fryb
|
furry homolog b (Drosophila) |
| chr20_+_16173618 | 0.80 |
ENSDART00000192109
ENSDART00000104112 ENSDART00000129633 |
zyg11
|
zyg-11 homolog (C. elegans) |
| chr2_-_23479714 | 0.79 |
ENSDART00000167291
|
si:ch211-14p21.3
|
si:ch211-14p21.3 |
| chr10_+_8629275 | 0.78 |
ENSDART00000129643
|
aplnrb
|
apelin receptor b |
| chr16_+_46294337 | 0.78 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr7_+_49664174 | 0.78 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr24_+_35564668 | 0.77 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr9_+_1654284 | 0.77 |
ENSDART00000062854
|
nfe2l2a
|
nuclear factor, erythroid 2-like 2a |
| chr13_-_23007813 | 0.77 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr1_-_51157454 | 0.76 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr5_+_483965 | 0.76 |
ENSDART00000150007
|
tek
|
TEK tyrosine kinase, endothelial |
| chr6_-_59942335 | 0.76 |
ENSDART00000168416
|
fbxl3b
|
F-box and leucine-rich repeat protein 3b |
| chr5_-_52784152 | 0.76 |
ENSDART00000169307
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr8_+_2456854 | 0.75 |
ENSDART00000133938
ENSDART00000002764 |
polb
|
polymerase (DNA directed), beta |
| chr18_-_3166726 | 0.74 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr23_-_44848961 | 0.74 |
ENSDART00000136839
|
wu:fb72h05
|
wu:fb72h05 |
| chr16_+_50089417 | 0.74 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr6_+_9421279 | 0.74 |
ENSDART00000161036
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr22_+_1006573 | 0.74 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr14_-_33454595 | 0.74 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr22_-_17729778 | 0.74 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr14_+_30910114 | 0.73 |
ENSDART00000187166
ENSDART00000078187 |
foxo4
|
forkhead box O4 |
| chr12_+_695619 | 0.73 |
ENSDART00000161691
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr11_-_236984 | 0.73 |
ENSDART00000170778
|
dusp7
|
dual specificity phosphatase 7 |
| chr25_+_37443194 | 0.73 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr20_+_13894123 | 0.73 |
ENSDART00000007744
|
slc30a1a
|
solute carrier family 30 (zinc transporter), member 1a |
| chr19_-_9472893 | 0.73 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr7_-_16596938 | 0.73 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr15_-_21837207 | 0.72 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr19_-_44091405 | 0.71 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr20_+_33294428 | 0.71 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr3_-_33417826 | 0.71 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr15_-_20468302 | 0.70 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr23_-_40817792 | 0.69 |
ENSDART00000136343
|
si:dkeyp-27c8.1
|
si:dkeyp-27c8.1 |
| chr24_+_42131564 | 0.69 |
ENSDART00000153854
|
wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr3_+_36284986 | 0.69 |
ENSDART00000059533
|
wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr1_-_411331 | 0.68 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr4_+_57093908 | 0.66 |
ENSDART00000170198
|
si:ch211-238e22.5
|
si:ch211-238e22.5 |
| chr5_-_2689753 | 0.66 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr22_-_607812 | 0.66 |
ENSDART00000145983
|
cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr3_+_5575313 | 0.65 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr22_-_367569 | 0.65 |
ENSDART00000041895
|
ssu72
|
SSU72 homolog, RNA polymerase II CTD phosphatase |
| chr19_+_48251273 | 0.64 |
ENSDART00000157424
|
CU693379.1
|
|
| chr3_+_22079219 | 0.64 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr22_+_26703026 | 0.64 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr18_+_36625490 | 0.63 |
ENSDART00000148032
|
clasrp
|
CLK4-associating serine/arginine rich protein |
| chr13_-_45523026 | 0.63 |
ENSDART00000020663
|
rhd
|
Rh blood group, D antigen |
| chr2_+_24177006 | 0.62 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr1_-_39943596 | 0.62 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr25_-_37331513 | 0.61 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr15_-_5742531 | 0.61 |
ENSDART00000045985
|
phkg1a
|
phosphorylase kinase, gamma 1a (muscle) |
| chr8_+_1187928 | 0.61 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr4_+_1600034 | 0.61 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr17_-_53329704 | 0.61 |
ENSDART00000193895
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr5_-_1047504 | 0.59 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr15_-_47857687 | 0.59 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr19_+_156757 | 0.59 |
ENSDART00000167717
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr20_-_16156419 | 0.58 |
ENSDART00000037420
|
ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr14_-_6225336 | 0.58 |
ENSDART00000111681
|
hdx
|
highly divergent homeobox |
| chr20_-_54381034 | 0.58 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr22_-_5441893 | 0.58 |
ENSDART00000161421
|
zgc:194627
|
zgc:194627 |
| chr6_-_54078623 | 0.58 |
ENSDART00000154076
|
hyal1
|
hyaluronoglucosaminidase 1 |
| chr15_+_19293744 | 0.57 |
ENSDART00000184994
ENSDART00000123815 |
jam3a
|
junctional adhesion molecule 3a |
| chr25_-_36492779 | 0.57 |
ENSDART00000042271
|
irx3b
|
iroquois homeobox 3b |
| chr12_-_48671612 | 0.57 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr25_+_17313568 | 0.57 |
ENSDART00000125459
|
cnot1
|
CCR4-NOT transcription complex, subunit 1 |
| chr20_+_18740518 | 0.56 |
ENSDART00000142196
|
fam167ab
|
family with sequence similarity 167, member Ab |
| chr9_+_993477 | 0.55 |
ENSDART00000182045
|
CABZ01066921.1
|
|
| chr7_-_66877058 | 0.55 |
ENSDART00000155954
|
adma
|
adrenomedullin a |
| chr18_+_36770166 | 0.55 |
ENSDART00000078151
|
fosb
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr1_+_54908895 | 0.55 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr4_-_9852318 | 0.55 |
ENSDART00000080702
|
glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr18_-_22414565 | 0.54 |
ENSDART00000079158
|
tmem170a
|
transmembrane protein 170A |
| chr9_-_8454060 | 0.54 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr2_+_24177190 | 0.53 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr23_+_14217508 | 0.53 |
ENSDART00000143618
|
birc7
|
baculoviral IAP repeat containing 7 |
| chr19_-_7450796 | 0.52 |
ENSDART00000104750
|
mllt11
|
MLLT11, transcription factor 7 cofactor |
| chr17_+_53418445 | 0.52 |
ENSDART00000097631
|
slc9a1b
|
solute carrier family 9 member A1b |
| chr4_-_23839789 | 0.52 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr18_-_3464222 | 0.51 |
ENSDART00000166980
|
myo7aa
|
myosin VIIAa |
| chr5_+_22969651 | 0.51 |
ENSDART00000089992
ENSDART00000145477 |
hmgn7
|
high mobility group nucleosomal binding domain 7 |
| chr21_-_308852 | 0.51 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr18_+_507618 | 0.51 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr16_-_52733384 | 0.50 |
ENSDART00000147236
ENSDART00000056101 |
azin1a
|
antizyme inhibitor 1a |
| chr11_-_497854 | 0.50 |
ENSDART00000104520
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr13_+_4405282 | 0.50 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr25_+_150570 | 0.49 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr18_-_7810214 | 0.49 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr2_-_42065069 | 0.49 |
ENSDART00000140188
|
cspp1b
|
centrosome and spindle pole associated protein 1b |
| chr16_-_8613499 | 0.49 |
ENSDART00000189189
|
cobl
|
cordon-bleu WH2 repeat protein |
| chr11_+_40831620 | 0.49 |
ENSDART00000160023
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.6 | 2.9 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.6 | 4.0 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.6 | 1.7 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.5 | 1.4 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.3 | 1.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.3 | 2.3 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.3 | 0.9 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.3 | 0.8 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.3 | 1.9 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.3 | 0.8 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.3 | 1.9 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.3 | 1.3 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.3 | 1.8 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.2 | 0.7 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.2 | 4.1 | GO:0001539 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 0.6 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.2 | 0.8 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 0.8 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.2 | 0.6 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.2 | 0.8 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903589) |
| 0.2 | 1.5 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.2 | 0.9 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.2 | 0.5 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.2 | 0.9 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.2 | 2.1 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 1.1 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.2 | 1.9 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.2 | 1.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.1 | 2.0 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.9 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.1 | 1.9 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.9 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.5 | GO:1902590 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.1 | 0.7 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 1.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 | 0.4 | GO:0097037 | heme export(GO:0097037) |
| 0.1 | 2.0 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.3 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.1 | 1.7 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 3.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.1 | 1.0 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 1.0 | GO:0034694 | response to prostaglandin(GO:0034694) |
| 0.1 | 0.1 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.8 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.7 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.1 | 0.3 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.6 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.1 | 0.7 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 1.0 | GO:0030431 | sleep(GO:0030431) |
| 0.1 | 0.7 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 1.6 | GO:1901663 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 0.2 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.3 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.1 | 0.4 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.4 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.1 | 0.5 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.4 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.1 | 0.2 | GO:1905132 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.4 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 0.4 | GO:0008206 | bile acid biosynthetic process(GO:0006699) bile acid metabolic process(GO:0008206) |
| 0.1 | 1.0 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.7 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 1.2 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 0.5 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 1.1 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.1 | 0.4 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 0.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.3 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 1.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.2 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 1.4 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
| 0.0 | 0.2 | GO:0036268 | swimming(GO:0036268) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.8 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.6 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 2.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.9 | GO:0060232 | delamination(GO:0060232) |
| 0.0 | 0.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.0 | 0.1 | GO:0090183 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.8 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 1.0 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.6 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0014814 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 1.0 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.6 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.7 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.2 | GO:2000765 | regulation of cytoplasmic translation(GO:2000765) negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.2 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.4 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 0.2 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.4 | GO:1903286 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.3 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.6 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 2.7 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 2.8 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.0 | 0.1 | GO:1902259 | regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.2 | GO:0061384 | trabecula morphogenesis(GO:0061383) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.8 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.8 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 1.4 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.7 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 0.5 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.5 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.4 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.5 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.2 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:1901315 | regulation of histone ubiquitination(GO:0033182) negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 1.0 | GO:0007596 | blood coagulation(GO:0007596) |
| 0.0 | 0.3 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.4 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.1 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.4 | GO:0042129 | regulation of T cell proliferation(GO:0042129) |
| 0.0 | 0.3 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.4 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.3 | 2.0 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.3 | 1.0 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.2 | 2.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 1.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.2 | 2.0 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.6 | GO:1990923 | PET complex(GO:1990923) |
| 0.1 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.7 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 1.8 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.5 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.4 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 0.7 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 3.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.1 | 1.0 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.6 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.6 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.6 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.5 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 4.9 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 1.2 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 1.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.7 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 5.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 1.4 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.8 | 4.0 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.6 | 1.7 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.5 | 4.4 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.5 | 2.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.5 | 1.4 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.4 | 2.9 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.2 | 1.8 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.2 | 1.3 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.2 | 1.3 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.2 | 1.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 1.6 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.2 | 0.8 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.2 | 0.8 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.2 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.8 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 2.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.6 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.1 | 0.9 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.7 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.9 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.8 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 3.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.6 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.1 | 0.7 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 1.6 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 2.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 2.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.1 | 0.6 | GO:0030331 | estrogen receptor binding(GO:0030331) retinoic acid receptor binding(GO:0042974) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 2.1 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 1.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.3 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.3 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 4.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.6 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.9 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.2 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 1.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 2.0 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 1.0 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 0.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.7 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.6 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 1.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 1.6 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 2.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.6 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.1 | GO:1904121 | phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.2 | GO:0034061 | DNA polymerase activity(GO:0034061) |
| 0.0 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.4 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.3 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 1.6 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.7 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.3 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.8 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0022842 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 1.0 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 2.3 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.4 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 1.8 | GO:0042802 | identical protein binding(GO:0042802) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 2.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 2.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 2.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID LKB1 PATHWAY | LKB1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.0 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.2 | 2.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.2 | 1.7 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.6 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.1 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 0.7 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 2.8 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 3.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 0.6 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 0.9 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.7 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 2.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 1.7 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.7 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.8 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |