Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
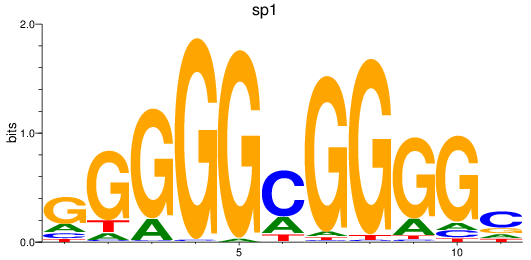
Results for sp1
Z-value: 2.23
Transcription factors associated with sp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp1
|
ENSDARG00000088347 | sp1 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp1 | dr11_v1_chr11_-_27821_27903 | 0.96 | 3.5e-10 | Click! |
Activity profile of sp1 motif
Sorted Z-values of sp1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_17860307 | 10.99 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr8_-_410728 | 9.55 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr18_-_127558 | 8.96 |
ENSDART00000149556
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_-_410199 | 7.29 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr18_-_127873 | 6.80 |
ENSDART00000148490
|
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr8_-_20230559 | 6.22 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr7_-_19332293 | 6.06 |
ENSDART00000169668
ENSDART00000137575 ENSDART00000090406 |
dock11
|
dedicator of cytokinesis 11 |
| chr3_+_51684963 | 6.01 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr16_-_7793457 | 5.86 |
ENSDART00000113483
|
trim71
|
tripartite motif containing 71, E3 ubiquitin protein ligase |
| chr17_-_23709347 | 5.80 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr2_+_35603637 | 5.78 |
ENSDART00000147278
|
plk3
|
polo-like kinase 3 (Drosophila) |
| chr7_+_38349667 | 5.75 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr6_+_3334392 | 5.67 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr20_-_182841 | 5.64 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr17_+_25856671 | 5.63 |
ENSDART00000064817
|
wapla
|
WAPL cohesin release factor a |
| chr25_+_3104959 | 5.62 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr5_-_54395488 | 5.40 |
ENSDART00000160781
|
zmynd19
|
zinc finger, MYND-type containing 19 |
| chr23_-_31645760 | 5.36 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr12_-_35505610 | 5.32 |
ENSDART00000105518
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr8_-_20230802 | 5.29 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr7_-_48251234 | 5.26 |
ENSDART00000024062
ENSDART00000098904 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr19_+_26340736 | 5.08 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| chr19_-_867071 | 5.08 |
ENSDART00000122257
|
eomesa
|
eomesodermin homolog a |
| chr6_+_27991943 | 5.06 |
ENSDART00000143974
ENSDART00000141354 ENSDART00000088914 ENSDART00000139367 |
amotl2a
|
angiomotin like 2a |
| chr6_+_32326074 | 4.99 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr5_-_13086616 | 4.98 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr23_-_45955177 | 4.92 |
ENSDART00000165963
ENSDART00000186649 ENSDART00000185773 |
LO017850.1
|
|
| chr12_+_46634736 | 4.79 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr23_+_2740741 | 4.75 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr6_-_1820606 | 4.75 |
ENSDART00000183228
|
FO834857.1
|
|
| chr10_-_2942900 | 4.68 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr7_+_44802353 | 4.67 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr19_+_30867845 | 4.67 |
ENSDART00000047461
|
mfsd2ab
|
major facilitator superfamily domain containing 2ab |
| chr17_-_50071748 | 4.66 |
ENSDART00000075188
|
zgc:113886
|
zgc:113886 |
| chr19_-_11031145 | 4.59 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr7_+_1473929 | 4.56 |
ENSDART00000050687
|
lpcat4
|
lysophosphatidylcholine acyltransferase 4 |
| chr19_-_2029777 | 4.54 |
ENSDART00000128639
|
CABZ01071939.1
|
|
| chr15_+_29025090 | 4.41 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr8_-_38022298 | 4.40 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr13_+_45582391 | 4.39 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr17_+_12075805 | 4.36 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr13_+_1100197 | 4.35 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr17_+_14965570 | 4.34 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr16_-_29387215 | 4.28 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr22_+_336256 | 4.26 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr2_+_23006792 | 4.22 |
ENSDART00000027782
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr4_-_4256300 | 4.22 |
ENSDART00000103319
ENSDART00000150279 |
cd9b
|
CD9 molecule b |
| chr20_+_39250673 | 4.20 |
ENSDART00000153003
|
reps1
|
RALBP1 associated Eps domain containing 1 |
| chr25_+_22319940 | 4.17 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr10_-_244745 | 4.17 |
ENSDART00000136551
|
klhl35
|
kelch-like family member 35 |
| chr15_+_29024895 | 4.17 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr7_+_7630409 | 4.15 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr21_-_43398457 | 4.13 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr21_-_30293224 | 4.12 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr12_-_37449396 | 4.05 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr14_-_15763648 | 4.05 |
ENSDART00000160712
|
dusp1
|
dual specificity phosphatase 1 |
| chr20_+_51061695 | 4.04 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr3_-_15470944 | 4.03 |
ENSDART00000185302
|
spns1
|
spinster homolog 1 (Drosophila) |
| chr4_-_1720648 | 4.01 |
ENSDART00000103484
|
gas2l3
|
growth arrest-specific 2 like 3 |
| chr15_-_25435085 | 4.01 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr23_-_10175898 | 4.00 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr6_+_12503849 | 3.94 |
ENSDART00000149529
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr3_-_15131438 | 3.94 |
ENSDART00000131720
|
xpo6
|
exportin 6 |
| chr2_+_25658112 | 3.92 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr7_-_4125021 | 3.91 |
ENSDART00000167182
ENSDART00000173696 |
zgc:55733
|
zgc:55733 |
| chr3_-_32362872 | 3.90 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr23_+_2825940 | 3.82 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr9_+_2452672 | 3.81 |
ENSDART00000193993
|
chn1
|
chimerin 1 |
| chr19_+_20201254 | 3.73 |
ENSDART00000010140
|
igf2bp3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr2_-_47620806 | 3.69 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr5_-_54714525 | 3.68 |
ENSDART00000150138
ENSDART00000150070 |
ccnb1
|
cyclin B1 |
| chr3_+_29082267 | 3.67 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr15_+_11644866 | 3.65 |
ENSDART00000188716
|
slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr10_+_39199547 | 3.63 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr5_-_54714789 | 3.62 |
ENSDART00000063357
|
ccnb1
|
cyclin B1 |
| chr23_+_35426404 | 3.60 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr2_-_52550135 | 3.49 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr21_-_43398122 | 3.46 |
ENSDART00000050533
|
ccni2
|
cyclin I family, member 2 |
| chr20_+_26940178 | 3.45 |
ENSDART00000190888
|
cdca4
|
cell division cycle associated 4 |
| chr15_+_46853252 | 3.45 |
ENSDART00000186040
|
zgc:153039
|
zgc:153039 |
| chr4_-_77551860 | 3.45 |
ENSDART00000188176
|
AL935186.6
|
|
| chr22_+_883678 | 3.44 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr16_-_26820634 | 3.43 |
ENSDART00000111156
|
pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr17_+_51746830 | 3.42 |
ENSDART00000184230
|
odc1
|
ornithine decarboxylase 1 |
| chr8_+_11687254 | 3.42 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr2_+_44512324 | 3.41 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr10_-_25217347 | 3.36 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr2_+_25657958 | 3.35 |
ENSDART00000161407
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr2_-_58075414 | 3.35 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr11_-_43226255 | 3.34 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr6_+_41503854 | 3.34 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr17_-_2578026 | 3.31 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr21_+_34122801 | 3.29 |
ENSDART00000182627
|
hmgb3b
|
high mobility group box 3b |
| chr16_-_8120203 | 3.27 |
ENSDART00000193430
|
snrka
|
SNF related kinase a |
| chr25_+_7492663 | 3.27 |
ENSDART00000166496
|
cat
|
catalase |
| chr14_+_32926385 | 3.25 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr9_-_46415847 | 3.22 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr7_-_45852270 | 3.22 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr9_-_11263228 | 3.21 |
ENSDART00000113847
|
chpfa
|
chondroitin polymerizing factor a |
| chr9_-_34269066 | 3.20 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr3_-_34547000 | 3.18 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr21_-_39024754 | 3.18 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr19_+_28291062 | 3.15 |
ENSDART00000163382
|
lpcat1
|
lysophosphatidylcholine acyltransferase 1 |
| chr18_+_3243292 | 3.14 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr17_-_2584423 | 3.13 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr20_-_28698172 | 3.12 |
ENSDART00000190635
|
sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr15_+_47582207 | 3.12 |
ENSDART00000159388
|
CABZ01087566.1
|
|
| chr10_-_22150419 | 3.12 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr22_+_835728 | 3.11 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr9_-_32343673 | 3.10 |
ENSDART00000078499
|
rftn2
|
raftlin family member 2 |
| chr14_+_30291611 | 3.10 |
ENSDART00000173004
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr23_-_32236122 | 3.10 |
ENSDART00000103343
|
grasp
|
GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein |
| chr3_+_15776446 | 3.08 |
ENSDART00000146651
|
znf652
|
zinc finger protein 652 |
| chr1_+_45707219 | 3.08 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr1_+_44196236 | 3.08 |
ENSDART00000179560
ENSDART00000166324 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr2_+_59015878 | 3.07 |
ENSDART00000148816
ENSDART00000122795 |
si:ch1073-391i24.1
|
si:ch1073-391i24.1 |
| chr13_+_49727333 | 3.05 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr24_+_35387517 | 3.04 |
ENSDART00000058571
|
snai2
|
snail family zinc finger 2 |
| chr3_-_30909487 | 3.04 |
ENSDART00000025046
|
ppp1caa
|
protein phosphatase 1, catalytic subunit, alpha isozyme a |
| chr18_-_3527686 | 3.02 |
ENSDART00000169049
|
capn5a
|
calpain 5a |
| chr17_+_4368859 | 3.02 |
ENSDART00000055385
|
crls1
|
cardiolipin synthase 1 |
| chr20_+_46741074 | 3.01 |
ENSDART00000145294
|
si:ch211-57i17.1
|
si:ch211-57i17.1 |
| chr24_+_24086249 | 3.00 |
ENSDART00000002953
|
lipib
|
lipase, member Ib |
| chr23_+_36306539 | 2.96 |
ENSDART00000053267
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr13_+_421231 | 2.95 |
ENSDART00000188212
ENSDART00000017854 |
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr7_+_67325933 | 2.94 |
ENSDART00000170575
ENSDART00000183342 |
nfat5b
|
nuclear factor of activated T cells 5b |
| chr15_-_25094026 | 2.94 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr19_-_6193448 | 2.94 |
ENSDART00000151405
|
erf
|
Ets2 repressor factor |
| chr6_+_21005725 | 2.93 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr6_+_38626684 | 2.93 |
ENSDART00000086533
|
atp10a
|
ATPase phospholipid transporting 10A |
| chr16_+_25259313 | 2.92 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr11_+_31324335 | 2.92 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr13_-_24218795 | 2.90 |
ENSDART00000136217
|
galnt2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 |
| chr3_+_43102010 | 2.89 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr20_+_4157815 | 2.87 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr19_+_14352332 | 2.83 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr7_+_10701938 | 2.83 |
ENSDART00000158162
|
arnt2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr1_-_40102836 | 2.82 |
ENSDART00000147317
|
cntf
|
ciliary neurotrophic factor |
| chr8_-_4618653 | 2.82 |
ENSDART00000025535
|
sept5a
|
septin 5a |
| chr20_-_53949798 | 2.82 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr3_-_61494840 | 2.81 |
ENSDART00000101957
|
baiap2l1b
|
BAI1-associated protein 2-like 1b |
| chr16_-_41667101 | 2.80 |
ENSDART00000084528
|
atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr6_-_7776612 | 2.80 |
ENSDART00000190269
|
myh9a
|
myosin, heavy chain 9a, non-muscle |
| chr22_+_11756040 | 2.79 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr7_+_24881680 | 2.77 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr4_-_77563411 | 2.76 |
ENSDART00000186841
|
AL935186.8
|
|
| chr12_-_25380028 | 2.76 |
ENSDART00000142674
|
zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr22_-_17688868 | 2.74 |
ENSDART00000012336
ENSDART00000147070 |
tjp3
|
tight junction protein 3 |
| chr3_-_6417328 | 2.74 |
ENSDART00000160979
|
jpt1b
|
Jupiter microtubule associated homolog 1b |
| chr13_+_11829072 | 2.73 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr19_-_23227582 | 2.71 |
ENSDART00000042172
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr3_+_34986837 | 2.71 |
ENSDART00000190341
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr15_-_45538773 | 2.71 |
ENSDART00000113494
|
MB21D2
|
Mab-21 domain containing 2 |
| chr22_-_34979139 | 2.71 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr15_+_12435975 | 2.70 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr15_+_40188076 | 2.70 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr18_+_27337994 | 2.69 |
ENSDART00000136172
|
si:dkey-29p10.4
|
si:dkey-29p10.4 |
| chr9_+_22780901 | 2.69 |
ENSDART00000110992
ENSDART00000143972 |
rif1
|
replication timing regulatory factor 1 |
| chr8_+_2656231 | 2.69 |
ENSDART00000160833
|
fam102aa
|
family with sequence similarity 102, member Aa |
| chr15_+_12436220 | 2.68 |
ENSDART00000169894
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr2_-_39675829 | 2.68 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr1_+_46493944 | 2.67 |
ENSDART00000114083
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr20_-_10487951 | 2.65 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr13_+_47821524 | 2.65 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr8_+_2656681 | 2.64 |
ENSDART00000185067
ENSDART00000165943 |
fam102aa
|
family with sequence similarity 102, member Aa |
| chr20_-_1635922 | 2.61 |
ENSDART00000181502
|
CR846082.1
|
|
| chr15_-_25093680 | 2.60 |
ENSDART00000062695
|
exo5
|
exonuclease 5 |
| chr3_-_36440705 | 2.60 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr10_-_35257458 | 2.60 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr12_-_4475890 | 2.58 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr12_+_14149686 | 2.58 |
ENSDART00000123741
|
kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr3_-_60589292 | 2.58 |
ENSDART00000157822
|
jmjd6
|
jumonji domain containing 6 |
| chr18_+_46151505 | 2.57 |
ENSDART00000015034
ENSDART00000141287 |
blvrb
|
biliverdin reductase B |
| chr18_+_38774584 | 2.57 |
ENSDART00000129750
|
fam214a
|
family with sequence similarity 214, member A |
| chr19_-_6193067 | 2.56 |
ENSDART00000092656
ENSDART00000140347 |
erf
|
Ets2 repressor factor |
| chr7_-_64971839 | 2.56 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr13_+_11828516 | 2.55 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr19_-_23249822 | 2.54 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr2_+_15100742 | 2.53 |
ENSDART00000027171
|
f3b
|
coagulation factor IIIb |
| chr23_-_36305874 | 2.51 |
ENSDART00000147598
ENSDART00000146986 ENSDART00000086985 ENSDART00000133259 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr22_-_20814450 | 2.51 |
ENSDART00000089076
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr11_-_12158412 | 2.51 |
ENSDART00000147670
|
npepps
|
aminopeptidase puromycin sensitive |
| chr22_+_5574952 | 2.51 |
ENSDART00000171774
|
zgc:171566
|
zgc:171566 |
| chr8_-_26709959 | 2.50 |
ENSDART00000135215
|
tmem51a
|
transmembrane protein 51a |
| chr24_+_24086491 | 2.50 |
ENSDART00000145092
|
lipib
|
lipase, member Ib |
| chr2_+_25560556 | 2.50 |
ENSDART00000133623
|
pld1a
|
phospholipase D1a |
| chr21_-_2042037 | 2.49 |
ENSDART00000171131
ENSDART00000160144 |
add1
|
adducin 1 (alpha) |
| chr12_-_4249000 | 2.49 |
ENSDART00000059298
|
zgc:92313
|
zgc:92313 |
| chr22_-_20924564 | 2.48 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr23_-_36306337 | 2.48 |
ENSDART00000142760
ENSDART00000136929 ENSDART00000143340 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr7_+_34794829 | 2.48 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr6_+_48206535 | 2.47 |
ENSDART00000075172
|
cttnbp2nla
|
CTTNBP2 N-terminal like a |
| chr16_-_22585289 | 2.47 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr5_+_4533244 | 2.47 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr15_-_31177324 | 2.47 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr4_-_5795309 | 2.45 |
ENSDART00000039987
|
pgm3
|
phosphoglucomutase 3 |
| chr9_+_22677503 | 2.45 |
ENSDART00000131429
ENSDART00000080005 ENSDART00000101756 ENSDART00000138148 |
itgb5
|
integrin, beta 5 |
| chr14_-_31694274 | 2.44 |
ENSDART00000173353
|
map7d3
|
MAP7 domain containing 3 |
| chr21_-_37027252 | 2.43 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr3_-_21118969 | 2.43 |
ENSDART00000129016
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 2.7 | 11.0 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 2.2 | 6.6 | GO:0034035 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 1.9 | 5.7 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 1.9 | 5.6 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.7 | 5.1 | GO:0060571 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 1.4 | 5.8 | GO:0048308 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 1.4 | 4.2 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 1.3 | 5.3 | GO:0042306 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 1.3 | 3.8 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 1.2 | 3.7 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 1.2 | 5.9 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 1.1 | 3.4 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 1.1 | 8.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 1.1 | 3.3 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 1.1 | 5.3 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 1.0 | 4.0 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 1.0 | 3.9 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 1.0 | 3.8 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.9 | 4.7 | GO:0045056 | transcytosis(GO:0045056) |
| 0.9 | 6.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.9 | 3.4 | GO:1904182 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.8 | 3.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.8 | 4.2 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.8 | 4.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.8 | 2.5 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.8 | 2.4 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.8 | 7.3 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.8 | 2.4 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.8 | 6.3 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.8 | 2.3 | GO:1903792 | negative regulation of anion transport(GO:1903792) |
| 0.7 | 2.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.7 | 4.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.7 | 11.5 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.7 | 4.0 | GO:1900744 | regulation of p38MAPK cascade(GO:1900744) |
| 0.7 | 2.0 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.6 | 4.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.6 | 1.9 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.6 | 1.9 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.6 | 5.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.6 | 6.4 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.6 | 5.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.6 | 2.5 | GO:2000677 | histone H3-K79 methylation(GO:0034729) regulation of transcription regulatory region DNA binding(GO:2000677) |
| 0.6 | 2.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.6 | 2.5 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.6 | 1.9 | GO:1990869 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.6 | 2.4 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.6 | 4.8 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.6 | 2.3 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.6 | 3.5 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.6 | 1.7 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.6 | 1.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.6 | 4.0 | GO:0021534 | cell proliferation in hindbrain(GO:0021534) |
| 0.6 | 2.8 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.6 | 2.8 | GO:0061439 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.6 | 2.2 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.5 | 4.4 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.5 | 3.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.5 | 3.8 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.5 | 1.6 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.5 | 3.2 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.5 | 3.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.5 | 3.1 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.5 | 1.5 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.5 | 1.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.5 | 5.0 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.4 | 1.3 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.4 | 3.1 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.4 | 6.6 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.4 | 1.8 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.4 | 3.4 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 | 1.7 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.4 | 1.7 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.4 | 0.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.4 | 2.0 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.4 | 1.9 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.4 | 1.5 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.4 | 2.9 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.4 | 2.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.4 | 4.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.4 | 3.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.4 | 1.8 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.4 | 0.7 | GO:0032211 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.3 | 1.4 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.3 | 1.0 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.3 | 2.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 5.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.3 | 1.3 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.3 | 1.6 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.3 | 4.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 1.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.3 | 1.5 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.3 | 1.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.3 | 2.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.3 | 1.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.3 | 1.7 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.3 | 6.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.3 | 1.4 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.3 | 0.8 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.3 | 2.8 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.3 | 5.8 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.3 | 2.7 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.3 | 1.6 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.3 | 3.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.3 | 1.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.3 | 2.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.3 | 0.8 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.3 | 2.0 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.2 | 2.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.9 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 0.7 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) glutamate receptor clustering(GO:0097688) |
| 0.2 | 0.5 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.2 | 1.7 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 3.5 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.2 | 1.6 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.2 | 2.0 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.2 | 2.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 0.7 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 2.0 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.2 | 1.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 0.9 | GO:0048011 | neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.2 | 1.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.2 | 1.7 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 5.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.2 | 1.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 6.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.2 | 0.8 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.2 | 3.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.2 | 1.8 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.2 | 1.0 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.2 | 1.8 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.2 | 2.0 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.2 | 0.6 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.2 | 0.6 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.2 | 0.6 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 2.0 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.3 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 3.3 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.2 | 0.7 | GO:0048521 | negative regulation of behavior(GO:0048521) negative regulation of feeding behavior(GO:2000252) |
| 0.2 | 0.9 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.2 | 1.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.2 | 0.9 | GO:0048885 | neuromast deposition(GO:0048885) |
| 0.2 | 0.7 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.2 | 1.8 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.2 | 1.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.2 | 2.4 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 2.1 | GO:0031937 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 0.8 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.2 | 1.3 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.2 | 0.5 | GO:1900180 | regulation of protein localization to nucleus(GO:1900180) |
| 0.2 | 1.7 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 2.0 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.2 | 2.6 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 0.7 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 1.6 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 1.7 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 4.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 1.5 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 1.8 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.6 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 6.0 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 0.8 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.1 | 0.7 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.5 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 0.8 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 2.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 2.1 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 8.2 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 1.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.1 | 4.7 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.1 | 3.9 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 1.0 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 1.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.6 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 1.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 1.4 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.6 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.7 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 2.8 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 1.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.1 | 9.0 | GO:0048916 | posterior lateral line development(GO:0048916) |
| 0.1 | 0.8 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.1 | 0.9 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 1.6 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.7 | GO:0003272 | endocardial cushion formation(GO:0003272) |
| 0.1 | 5.3 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.1 | 1.8 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.1 | 1.9 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.1 | 1.3 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 1.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.1 | 1.1 | GO:0032231 | regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 2.5 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.1 | 1.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 6.1 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 0.9 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.3 | GO:0044275 | cellular carbohydrate catabolic process(GO:0044275) |
| 0.1 | 3.7 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.1 | 1.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 2.6 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 9.6 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.1 | 0.2 | GO:0042560 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.1 | 0.8 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.4 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.9 | GO:0043931 | ossification involved in bone maturation(GO:0043931) bone maturation(GO:0070977) |
| 0.1 | 0.7 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 8.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 0.5 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.1 | 0.9 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 2.3 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.1 | 5.5 | GO:0051402 | neuron apoptotic process(GO:0051402) |
| 0.1 | 0.4 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.1 | 1.0 | GO:0003139 | secondary heart field specification(GO:0003139) |
| 0.1 | 0.5 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 6.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.1 | 1.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 2.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 5.3 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 2.6 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 0.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 2.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.1 | 0.2 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 1.4 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.1 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.1 | 2.3 | GO:0048538 | thymus development(GO:0048538) |
| 0.1 | 2.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 2.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 0.2 | GO:0001112 | DNA-templated transcriptional open complex formation(GO:0001112) transcriptional open complex formation at RNA polymerase II promoter(GO:0001113) protein-DNA complex remodeling(GO:0001120) |
| 0.1 | 0.3 | GO:0072506 | phosphate ion homeostasis(GO:0055062) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.1 | 6.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 13.5 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 4.5 | GO:0042552 | myelination(GO:0042552) |
| 0.1 | 1.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.1 | 1.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.5 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 6.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 1.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 1.2 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 0.9 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.1 | 0.9 | GO:0090114 | COPII-coated vesicle budding(GO:0090114) |
| 0.1 | 0.3 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.1 | 1.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.9 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 4.4 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.3 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 1.6 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.1 | 4.0 | GO:0044344 | fibroblast growth factor receptor signaling pathway(GO:0008543) cellular response to fibroblast growth factor stimulus(GO:0044344) response to fibroblast growth factor(GO:0071774) |
| 0.1 | 1.0 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.5 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 0.3 | GO:0071380 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.2 | GO:0014909 | smooth muscle cell migration(GO:0014909) |
| 0.1 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.7 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 1.6 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 2.3 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.1 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0003245 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 1.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 2.0 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.4 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 5.4 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 1.3 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 4.7 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 2.1 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 6.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.2 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 1.1 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 2.9 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.0 | 2.1 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 1.1 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.5 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 4.4 | GO:0016236 | macroautophagy(GO:0016236) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 1.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.8 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 1.9 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 2.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.2 | GO:1902742 | apoptotic process involved in morphogenesis(GO:0060561) apoptotic process involved in development(GO:1902742) |
| 0.0 | 0.3 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 0.0 | GO:0007612 | learning(GO:0007612) |
| 0.0 | 1.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.0 | 0.4 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.4 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.2 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0001757 | somite specification(GO:0001757) segment specification(GO:0007379) |
| 0.0 | 2.6 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.4 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 5.7 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.8 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:1903321 | negative regulation of protein ubiquitination(GO:0031397) negative regulation of protein modification by small protein conjugation or removal(GO:1903321) |
| 0.0 | 0.1 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 2.3 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.6 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 1.2 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.0 | 0.2 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.7 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.6 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 1.7 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.5 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) |
| 0.0 | 2.7 | GO:0006644 | phospholipid metabolic process(GO:0006644) |
| 0.0 | 2.1 | GO:0006399 | tRNA metabolic process(GO:0006399) |
| 0.0 | 0.2 | GO:0035778 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.0 | 0.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.0 | 0.3 | GO:1904892 | regulation of JAK-STAT cascade(GO:0046425) regulation of STAT cascade(GO:1904892) |
| 0.0 | 0.3 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.1 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.2 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.5 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.7 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.1 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.2 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.2 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 1.0 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 1.3 | 5.3 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.8 | 2.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 3.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.7 | 2.6 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.6 | 5.0 | GO:0035060 | brahma complex(GO:0035060) |
| 0.5 | 8.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 10.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 3.8 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.4 | 4.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.4 | 3.4 | GO:0001401 | mitochondrial sorting and assembly machinery complex(GO:0001401) |
| 0.4 | 1.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.4 | 2.7 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.4 | 5.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.3 | 2.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.3 | 1.0 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.3 | 2.3 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 1.3 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.3 | 2.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.3 | 1.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.3 | 2.9 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.3 | 2.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.3 | 1.4 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.3 | 0.8 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.3 | 1.0 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.2 | 1.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.2 | 0.7 | GO:1990879 | CST complex(GO:1990879) |
| 0.2 | 0.9 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 2.6 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 15.5 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 2.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.2 | 1.4 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 1.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 1.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 1.9 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.2 | 23.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.2 | 3.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.6 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.2 | 1.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 3.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 0.8 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 6.1 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 1.8 | GO:0030904 | retromer complex(GO:0030904) |
| 0.2 | 2.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.2 | 0.6 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 0.6 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.2 | 2.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 1.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 1.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 1.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 6.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 7.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 1.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 10.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.4 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 1.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 8.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.1 | 0.5 | GO:0005903 | brush border(GO:0005903) |
| 0.1 | 3.4 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 1.8 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 1.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 4.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.6 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 4.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 3.5 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.4 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.1 | 3.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.7 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 8.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 1.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 0.6 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 0.5 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 1.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.8 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 2.5 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.1 | 5.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.4 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 0.9 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.5 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.5 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 1.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.9 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 5.2 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 3.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.9 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 1.2 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.8 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 2.9 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.0 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 9.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 2.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.6 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 20.5 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 2.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.1 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 9.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 13.7 | GO:0012505 | endomembrane system(GO:0012505) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 1.8 | 5.5 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 1.4 | 5.7 | GO:0071253 | connexin binding(GO:0071253) |
| 1.4 | 4.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.3 | 3.9 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 1.1 | 7.4 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 1.0 | 11.5 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 1.0 | 13.7 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.9 | 3.4 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.8 | 2.5 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.7 | 2.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.7 | 2.7 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.7 | 10.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.6 | 3.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.6 | 2.5 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.6 | 5.6 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.6 | 5.9 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.6 | 2.3 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.6 | 1.7 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.6 | 1.7 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.5 | 4.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 3.8 | GO:0070728 | leucine binding(GO:0070728) |
| 0.5 | 2.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.5 | 5.3 | GO:0035925 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.5 | 2.0 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.5 | 1.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.5 | 5.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.5 | 2.8 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.5 | 1.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.4 | 2.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.4 | 2.2 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.4 | 5.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.4 | 1.8 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.4 | 1.3 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.4 | 2.0 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.4 | 7.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 1.9 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 3.7 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.4 | 1.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.4 | 3.3 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 2.9 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.4 | 3.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 2.5 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.4 | 1.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.3 | 3.0 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.3 | 1.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 1.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 1.5 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.3 | 2.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 1.2 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.3 | 9.2 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.3 | 2.4 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.3 | 0.9 | GO:0043621 | protein self-association(GO:0043621) |
| 0.3 | 1.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 5.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.3 | 8.2 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 0.5 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.3 | 1.3 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.3 | 1.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.2 | 5.7 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 0.7 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.2 | 1.9 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.2 | 0.7 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.2 | 1.9 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.2 | 3.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.2 | 1.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 2.5 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.2 | 10.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.2 | 0.7 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 1.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.2 | 5.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 0.8 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 1.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 2.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 2.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.2 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 3.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 3.1 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.2 | 7.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 2.3 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.2 | 0.8 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.2 | 1.3 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 4.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.6 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.2 | 8.0 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.2 | 1.7 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 2.2 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.2 | 0.6 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.1 | 1.0 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.8 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 4.0 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 3.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 4.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 1.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 4.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.0 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.1 | 3.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.6 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 1.1 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 0.6 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 5.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 2.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 1.5 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 2.8 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.8 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.5 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 2.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.1 | 0.4 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 3.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.5 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.1 | 1.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.1 | 2.7 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.1 | 1.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 8.2 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 1.3 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.1 | 4.6 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.1 | 1.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.5 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 1.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.1 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.6 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 1.4 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.1 | 1.6 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.1 | 2.9 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.1 | 3.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 3.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.8 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 1.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 10.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 2.4 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.1 | 1.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 0.9 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 1.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 2.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.2 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 3.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.1 | 0.8 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 4.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 2.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.7 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 1.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 5.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 13.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.1 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.8 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 1.4 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 7.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.3 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 1.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 9.8 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 11.1 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 7.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.8 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.2 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 11.3 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.4 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 11.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.3 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.3 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 1.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 5.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.4 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.8 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 1.1 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 4.5 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 3.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.3 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 18.2 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 0.6 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 4.0 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 1.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.3 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.4 | 8.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.4 | 17.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.3 | 3.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.3 | 2.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.3 | 9.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 3.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 3.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.3 | 9.0 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.3 | 1.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.2 | 2.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.2 | 4.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.2 | 8.3 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.2 | 5.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 4.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.2 | 3.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.2 | 3.8 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 3.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.2 | 3.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.2 | 1.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.2 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 10.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.2 | 2.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 3.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.2 | 0.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 10.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 4.3 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 2.9 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 0.6 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 1.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 0.7 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.4 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 4.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.0 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | ST GA13 PATHWAY | G alpha 13 Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 10.7 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 1.0 | 3.8 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.7 | 7.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.7 | 3.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.6 | 5.6 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.4 | 3.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.4 | 2.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.4 | 4.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 9.8 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.3 | 1.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.3 | 6.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.3 | 2.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.3 | 2.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 0.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.3 | 4.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 3.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.2 | 3.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 2.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 4.1 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 0.8 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 9.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.2 | 1.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.2 | 3.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 3.7 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.2 | 1.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 1.9 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.7 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 3.7 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 3.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 2.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.5 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.1 | 9.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 1.4 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 19.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 2.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 2.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.7 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 2.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 1.5 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 2.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 0.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.1 | 6.0 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.1 | 6.0 | REACTOME DIABETES PATHWAYS | Genes involved in Diabetes pathways |
| 0.0 | 2.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 1.5 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.6 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |