Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
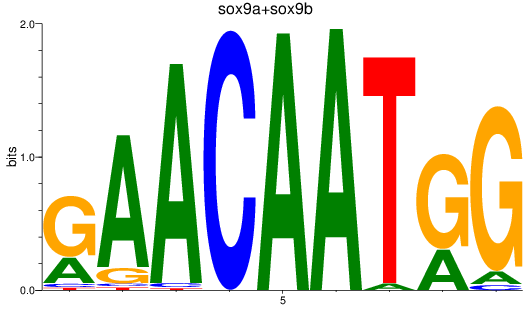
Results for sox9a+sox9b
Z-value: 1.78
Transcription factors associated with sox9a+sox9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox9a
|
ENSDARG00000003293 | SRY-box transcription factor 9a |
|
sox9b
|
ENSDARG00000043923 | SRY-box transcription factor 9b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox9a | dr11_v1_chr12_-_1951233_1951284 | -0.94 | 5.3e-09 | Click! |
| sox9b | dr11_v1_chr3_-_62527675_62527696 | 0.93 | 3.2e-08 | Click! |
Activity profile of sox9a+sox9b motif
Sorted Z-values of sox9a+sox9b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_32385989 | 7.15 |
ENSDART00000143716
ENSDART00000098850 |
lipg
|
lipase, endothelial |
| chr19_+_14109348 | 5.94 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr5_+_57924611 | 5.60 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr18_+_45571378 | 5.36 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr24_-_2381143 | 5.25 |
ENSDART00000144307
|
rreb1a
|
ras responsive element binding protein 1a |
| chr7_+_58751504 | 4.89 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr16_+_35535375 | 4.66 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr23_+_36308428 | 4.10 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr20_+_54304800 | 4.05 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr24_-_41797681 | 4.02 |
ENSDART00000169643
|
arhgap28
|
Rho GTPase activating protein 28 |
| chr16_+_35535171 | 4.02 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr10_+_15255012 | 4.01 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr10_+_15255198 | 3.99 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr22_-_10541372 | 3.88 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr20_+_54295213 | 3.88 |
ENSDART00000074085
|
zp2.3
|
zona pellucida glycoprotein 2, tandem duplicate 3 |
| chr16_+_26846495 | 3.84 |
ENSDART00000078124
|
trim35-29
|
tripartite motif containing 35-29 |
| chr20_+_54299419 | 3.82 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr20_+_54312970 | 3.81 |
ENSDART00000024598
ENSDART00000193172 |
zp2.5
|
zona pellucida glycoprotein 2, tandem duplicate 5 |
| chr15_+_29025090 | 3.76 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr20_+_54309148 | 3.73 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr18_-_7032227 | 3.61 |
ENSDART00000127138
|
calub
|
calumenin b |
| chr20_+_54290356 | 3.61 |
ENSDART00000173347
|
zp2.2
|
zona pellucida glycoprotein 2, tandem duplicate 2 |
| chr17_-_45125537 | 3.56 |
ENSDART00000113552
|
zgc:163014
|
zgc:163014 |
| chr8_-_410199 | 3.54 |
ENSDART00000091177
ENSDART00000122979 ENSDART00000151331 ENSDART00000151155 |
trim36
|
tripartite motif containing 36 |
| chr8_-_44611357 | 3.35 |
ENSDART00000063396
|
bag4
|
BCL2 associated athanogene 4 |
| chr24_-_19719240 | 3.32 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr3_+_32411343 | 3.20 |
ENSDART00000186287
ENSDART00000141793 |
rras
|
RAS related |
| chr22_-_22337382 | 3.10 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr12_+_30788912 | 3.10 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr3_+_32410746 | 3.06 |
ENSDART00000025496
|
rras
|
RAS related |
| chr19_+_7549854 | 3.02 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr22_+_15960514 | 3.00 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr11_-_44979281 | 2.90 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr5_-_56964547 | 2.82 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr13_-_35907768 | 2.80 |
ENSDART00000147522
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr22_+_14117078 | 2.79 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr13_-_36761379 | 2.78 |
ENSDART00000131534
ENSDART00000029824 |
map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr9_-_46415847 | 2.77 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr2_+_32846602 | 2.75 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr24_-_1985007 | 2.72 |
ENSDART00000189870
|
PARD3 (1 of many)
|
par-3 family cell polarity regulator |
| chr13_-_35908275 | 2.71 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr14_-_41478265 | 2.71 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr6_-_46742455 | 2.70 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr5_+_44846434 | 2.69 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr4_-_837768 | 2.68 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr7_+_22801465 | 2.67 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr11_+_41838801 | 2.66 |
ENSDART00000014871
|
akr7a3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr21_-_30082414 | 2.64 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr2_+_34967022 | 2.56 |
ENSDART00000134926
|
astn1
|
astrotactin 1 |
| chr13_-_6250317 | 2.56 |
ENSDART00000180416
|
tuba4l
|
tubulin, alpha 4 like |
| chr15_-_16076399 | 2.54 |
ENSDART00000135658
ENSDART00000133755 ENSDART00000080413 |
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr2_+_44512324 | 2.53 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr14_+_16287968 | 2.52 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr22_-_5171362 | 2.52 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr3_+_54047342 | 2.52 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr10_-_36691681 | 2.50 |
ENSDART00000122375
|
mrpl48
|
mitochondrial ribosomal protein L48 |
| chr22_+_15960005 | 2.50 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr5_+_44846280 | 2.48 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr16_-_54919260 | 2.45 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr22_+_15959844 | 2.42 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr9_-_14273652 | 2.40 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr5_+_58679071 | 2.39 |
ENSDART00000019561
|
zgc:171734
|
zgc:171734 |
| chr23_+_35426404 | 2.39 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr23_+_19590006 | 2.39 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr2_+_34967210 | 2.38 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr6_+_33931095 | 2.37 |
ENSDART00000191909
|
orc1
|
origin recognition complex, subunit 1 |
| chr15_-_31265375 | 2.37 |
ENSDART00000086592
|
vezf1b
|
vascular endothelial zinc finger 1b |
| chr5_-_22130937 | 2.36 |
ENSDART00000138606
|
las1l
|
LAS1-like, ribosome biogenesis factor |
| chr14_-_16810401 | 2.36 |
ENSDART00000158396
ENSDART00000170758 |
tcirg1b
|
T cell immune regulator 1, ATPase H+ transporting V0 subunit a3b |
| chr9_-_28937880 | 2.34 |
ENSDART00000132878
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr1_+_49878000 | 2.34 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr14_-_32876280 | 2.30 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr14_-_26704829 | 2.29 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr17_+_14965570 | 2.26 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr19_+_46113828 | 2.25 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr21_-_11054605 | 2.25 |
ENSDART00000191378
ENSDART00000084061 |
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr1_+_16574312 | 2.22 |
ENSDART00000187067
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr18_-_38270430 | 2.22 |
ENSDART00000139519
|
caprin1b
|
cell cycle associated protein 1b |
| chr3_+_53156813 | 2.20 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr12_+_48340133 | 2.20 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr9_+_24192370 | 2.18 |
ENSDART00000003482
|
stk17b
|
serine/threonine kinase 17b (apoptosis-inducing) |
| chr17_+_28005763 | 2.18 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr23_-_40536017 | 2.12 |
ENSDART00000153751
ENSDART00000140623 ENSDART00000133356 |
rnf146
|
ring finger protein 146 |
| chr7_+_19850889 | 2.12 |
ENSDART00000100782
|
mus81
|
MUS81 structure-specific endonuclease subunit |
| chr23_-_14403939 | 2.08 |
ENSDART00000090930
|
nkain4
|
sodium/potassium transporting ATPase interacting 4 |
| chr14_+_38786298 | 2.05 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr6_-_49547680 | 2.04 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr14_+_34490445 | 2.03 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr19_+_41479990 | 2.03 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr13_+_33368503 | 2.01 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr22_-_6420239 | 2.00 |
ENSDART00000148385
|
zgc:171699
|
zgc:171699 |
| chr9_-_34260214 | 1.99 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr6_-_11362871 | 1.99 |
ENSDART00000151125
|
pcnt
|
pericentrin |
| chr20_-_3319642 | 1.97 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr10_-_28028998 | 1.97 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr3_+_36127287 | 1.96 |
ENSDART00000058605
ENSDART00000182500 |
scpep1
|
serine carboxypeptidase 1 |
| chr16_+_35905031 | 1.95 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr1_+_46493944 | 1.94 |
ENSDART00000114083
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr3_-_26805455 | 1.93 |
ENSDART00000180648
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr2_-_27651674 | 1.92 |
ENSDART00000177402
|
tgs1
|
trimethylguanosine synthase 1 |
| chr7_-_39378903 | 1.92 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr11_-_27057572 | 1.91 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr10_+_15608326 | 1.90 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr15_+_11644866 | 1.90 |
ENSDART00000188716
|
slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr21_+_37513058 | 1.89 |
ENSDART00000141096
|
amot
|
angiomotin |
| chr7_-_39360325 | 1.88 |
ENSDART00000098033
ENSDART00000173695 ENSDART00000173466 ENSDART00000173734 |
ambra1a
|
autophagy/beclin-1 regulator 1a |
| chr14_+_16036139 | 1.88 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr20_+_13141408 | 1.88 |
ENSDART00000034098
|
dtl
|
denticleless E3 ubiquitin protein ligase homolog (Drosophila) |
| chr2_+_36608387 | 1.85 |
ENSDART00000159541
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr17_+_38602790 | 1.84 |
ENSDART00000062010
|
ccdc88c
|
coiled-coil domain containing 88C |
| chr20_+_26943072 | 1.84 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr1_+_218524 | 1.82 |
ENSDART00000109529
|
tmco3
|
transmembrane and coiled-coil domains 3 |
| chr6_-_12172424 | 1.82 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr9_-_22057658 | 1.81 |
ENSDART00000101944
|
crygmxl1
|
crystallin, gamma MX, like 1 |
| chr5_-_41124241 | 1.81 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr1_+_52632856 | 1.76 |
ENSDART00000011725
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr21_+_37513488 | 1.76 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr12_-_3053699 | 1.75 |
ENSDART00000139721
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr20_-_31238313 | 1.74 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr5_+_56277866 | 1.74 |
ENSDART00000170610
ENSDART00000028854 ENSDART00000148749 |
aatf
|
apoptosis antagonizing transcription factor |
| chr1_-_21714025 | 1.74 |
ENSDART00000129066
|
zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr21_-_11054876 | 1.72 |
ENSDART00000146576
|
nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like |
| chr12_+_33460794 | 1.70 |
ENSDART00000007053
ENSDART00000142716 |
narf
|
nuclear prelamin A recognition factor |
| chr24_-_2423791 | 1.69 |
ENSDART00000190402
|
rreb1a
|
ras responsive element binding protein 1a |
| chr10_-_15879569 | 1.69 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr23_-_29357764 | 1.68 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr24_-_22103585 | 1.68 |
ENSDART00000000070
|
dap
|
death-associated protein |
| chr25_+_35891342 | 1.68 |
ENSDART00000147093
|
lsm14aa
|
LSM14A mRNA processing body assembly factor a |
| chr20_-_40766387 | 1.66 |
ENSDART00000061173
|
hsdl1
|
hydroxysteroid dehydrogenase like 1 |
| chr15_+_25439106 | 1.66 |
ENSDART00000156252
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr25_+_18475032 | 1.65 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr3_+_17653784 | 1.64 |
ENSDART00000159984
ENSDART00000157682 ENSDART00000187937 |
kat2a
|
K(lysine) acetyltransferase 2A |
| chr23_+_9522942 | 1.63 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr2_-_21167652 | 1.63 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr20_+_6533260 | 1.62 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr8_+_42941555 | 1.61 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr12_+_16953415 | 1.59 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr11_+_18873619 | 1.59 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr14_-_24277805 | 1.58 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr23_+_38171186 | 1.57 |
ENSDART00000148188
|
zgc:112994
|
zgc:112994 |
| chr21_+_17301790 | 1.57 |
ENSDART00000145057
|
tsc1b
|
TSC complex subunit 1b |
| chr6_+_32326074 | 1.57 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr4_-_12040322 | 1.55 |
ENSDART00000150583
ENSDART00000102260 |
si:dkey-222f8.3
|
si:dkey-222f8.3 |
| chr17_+_33415319 | 1.54 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr10_-_28027490 | 1.53 |
ENSDART00000185445
|
ints2
|
integrator complex subunit 2 |
| chr4_+_5334202 | 1.53 |
ENSDART00000150409
|
apex1
|
APEX nuclease (multifunctional DNA repair enzyme) 1 |
| chr7_+_32897723 | 1.51 |
ENSDART00000146789
ENSDART00000140775 ENSDART00000142049 ENSDART00000145523 ENSDART00000135237 ENSDART00000133584 ENSDART00000140800 ENSDART00000137956 ENSDART00000075263 |
tssc4
|
tumor suppressing subtransferable candidate 4 |
| chr9_-_41401564 | 1.50 |
ENSDART00000059628
|
nab1b
|
NGFI-A binding protein 1b (EGR1 binding protein 1) |
| chr21_+_38089036 | 1.50 |
ENSDART00000147219
|
klf8
|
Kruppel-like factor 8 |
| chr15_+_23947932 | 1.50 |
ENSDART00000153951
|
myo18ab
|
myosin XVIIIAb |
| chr2_-_5135125 | 1.49 |
ENSDART00000164039
|
ptmab
|
prothymosin, alpha b |
| chr20_+_1272526 | 1.49 |
ENSDART00000008115
ENSDART00000133825 |
hsd3b2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr23_+_9522781 | 1.47 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr23_-_27050083 | 1.47 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr21_+_19368720 | 1.47 |
ENSDART00000187759
ENSDART00000185829 ENSDART00000158471 ENSDART00000168728 |
btc
|
betacellulin, epidermal growth factor family member |
| chr19_-_34011340 | 1.47 |
ENSDART00000172618
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr3_-_45778123 | 1.46 |
ENSDART00000146211
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr18_+_17493859 | 1.46 |
ENSDART00000090754
|
si:dkey-102f14.5
|
si:dkey-102f14.5 |
| chr6_-_15491579 | 1.46 |
ENSDART00000156439
|
st6gal2b
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2b |
| chr19_-_31042570 | 1.46 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr14_-_33978117 | 1.45 |
ENSDART00000128515
|
foxa
|
forkhead box A sequence |
| chr2_+_2967255 | 1.44 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr15_-_20839763 | 1.44 |
ENSDART00000141746
ENSDART00000182369 |
aldh3a2a
|
aldehyde dehydrogenase 3 family, member A2a |
| chr18_+_36066389 | 1.43 |
ENSDART00000059347
|
bckdha
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr22_+_4443689 | 1.43 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr6_-_37743724 | 1.42 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr14_+_31473866 | 1.42 |
ENSDART00000173088
|
ccdc160
|
coiled-coil domain containing 160 |
| chr18_-_38270077 | 1.41 |
ENSDART00000185546
|
caprin1b
|
cell cycle associated protein 1b |
| chr23_+_37482727 | 1.40 |
ENSDART00000162737
|
agmat
|
agmatine ureohydrolase (agmatinase) |
| chr9_-_34500197 | 1.40 |
ENSDART00000114043
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr7_-_19998723 | 1.39 |
ENSDART00000173458
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr3_-_49514874 | 1.39 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr24_-_11905911 | 1.39 |
ENSDART00000033621
|
tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr7_-_59514547 | 1.38 |
ENSDART00000168457
|
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr24_-_11076400 | 1.37 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr2_+_26240631 | 1.37 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr15_+_23722620 | 1.37 |
ENSDART00000011447
|
sae1
|
SUMO1 activating enzyme subunit 1 |
| chr17_-_33415740 | 1.37 |
ENSDART00000135218
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr24_+_26140855 | 1.36 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr18_-_45761868 | 1.36 |
ENSDART00000025423
|
cstf3
|
cleavage stimulation factor, 3' pre-RNA, subunit 3 |
| chr5_+_25762271 | 1.36 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr8_-_11834599 | 1.35 |
ENSDART00000190986
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr23_-_27235403 | 1.35 |
ENSDART00000134418
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr7_-_30624435 | 1.35 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr7_-_58098814 | 1.35 |
ENSDART00000147287
ENSDART00000043984 |
ank2b
|
ankyrin 2b, neuronal |
| chr7_-_40959667 | 1.32 |
ENSDART00000084070
|
rbm33a
|
RNA binding motif protein 33a |
| chr19_-_7321221 | 1.31 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr17_-_7028418 | 1.31 |
ENSDART00000188305
ENSDART00000187895 |
sash1b
|
SAM and SH3 domain containing 1b |
| chr22_-_20838011 | 1.31 |
ENSDART00000186056
|
dot1l
|
DOT1-like histone H3K79 methyltransferase |
| chr10_+_37173029 | 1.31 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr18_+_45666489 | 1.31 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr19_-_20446756 | 1.31 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr17_+_33415542 | 1.29 |
ENSDART00000183169
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr18_+_20034023 | 1.29 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr2_+_26303627 | 1.28 |
ENSDART00000040278
|
efna2a
|
ephrin-A2a |
| chr12_-_17592215 | 1.28 |
ENSDART00000134597
|
usp42
|
ubiquitin specific peptidase 42 |
| chr2_-_9696283 | 1.28 |
ENSDART00000165712
|
selenot1a
|
selenoprotein T, 1a |
| chr13_+_28580357 | 1.28 |
ENSDART00000007211
|
wbp1la
|
WW domain binding protein 1-like a |
| chr17_+_24597001 | 1.28 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr2_-_26596794 | 1.27 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox9a+sox9b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.9 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.8 | 3.3 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.7 | 2.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.7 | 2.0 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.6 | 3.1 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.6 | 8.6 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.6 | 1.8 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.5 | 1.6 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.5 | 4.0 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.5 | 2.4 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.5 | 1.4 | GO:0035666 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.5 | 2.3 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.5 | 2.3 | GO:0048909 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.5 | 3.6 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.4 | 8.8 | GO:0051098 | regulation of binding(GO:0051098) |
| 0.4 | 7.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.4 | 2.5 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.4 | 2.0 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.4 | 1.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.4 | 1.1 | GO:0072388 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.4 | 1.5 | GO:1902745 | skeletal muscle cell proliferation(GO:0014856) regulation of skeletal muscle cell proliferation(GO:0014857) positive regulation of lamellipodium organization(GO:1902745) |
| 0.4 | 1.1 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.3 | 1.4 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.3 | 3.1 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 1.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.3 | 1.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 2.5 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.3 | 0.9 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.3 | 2.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 0.9 | GO:0046833 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.3 | 1.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 2.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.3 | 1.1 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.2 | 1.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 2.4 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 1.9 | GO:0072425 | mitotic G2 DNA damage checkpoint(GO:0007095) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.2 | 1.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 0.7 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.2 | 0.6 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.2 | 1.7 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.2 | 1.0 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 2.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.2 | 1.0 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 2.2 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) nephric duct morphogenesis(GO:0072178) |
| 0.2 | 2.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.2 | 0.6 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.2 | 1.6 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 1.4 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.2 | 1.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.2 | 2.4 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 2.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 1.8 | GO:0021694 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 | 1.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.2 | 0.8 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.2 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.6 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.2 | 2.6 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.2 | 2.8 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 0.6 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.2 | 1.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 4.2 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 1.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 1.5 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.1 | 1.3 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 1.6 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 2.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.8 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.6 | GO:0099638 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 3.5 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.1 | 1.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.2 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.3 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 1.7 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 2.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.1 | 0.4 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.3 | GO:0050666 | regulation of sulfur amino acid metabolic process(GO:0031335) regulation of homocysteine metabolic process(GO:0050666) |
| 0.1 | 1.4 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 2.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.2 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.1 | 3.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 1.3 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 3.9 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 9.4 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 0.6 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 1.5 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.0 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 3.2 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.1 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.1 | 1.1 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 1.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.6 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.0 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 1.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.2 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.1 | 2.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 0.6 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.1 | 0.2 | GO:0003403 | optic vesicle formation(GO:0003403) |
| 0.1 | 1.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.6 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 0.7 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 | 0.5 | GO:2001271 | regulation of execution phase of apoptosis(GO:1900117) negative regulation of execution phase of apoptosis(GO:1900118) regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001270) negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 6.3 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.1 | 0.3 | GO:0051972 | regulation of telomerase activity(GO:0051972) |
| 0.1 | 0.5 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 1.3 | GO:0042953 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 0.3 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.8 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 2.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.7 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 4.4 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 1.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.7 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0030656 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 1.2 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 1.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 1.5 | GO:0060415 | muscle tissue morphogenesis(GO:0060415) |
| 0.0 | 0.8 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 4.7 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.7 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 1.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 2.5 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 1.1 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 1.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 3.8 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.6 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 2.5 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.7 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.0 | 3.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 1.4 | GO:0031123 | RNA 3'-end processing(GO:0031123) |
| 0.0 | 1.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.2 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 1.4 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.6 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 1.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 2.4 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 2.6 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.6 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 1.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.2 | GO:0006622 | protein targeting to lysosome(GO:0006622) protein retention in Golgi apparatus(GO:0045053) |
| 0.0 | 0.5 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 1.4 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 0.9 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.8 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.0 | 0.5 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.6 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 1.4 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.0 | 0.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.9 | GO:0034968 | histone lysine methylation(GO:0034968) |
| 0.0 | 0.5 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 5.1 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.5 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.1 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 0.6 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.3 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.2 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.9 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.6 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.8 | GO:0060048 | cardiac muscle contraction(GO:0060048) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 1.5 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 2.7 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.8 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.1 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0006693 | prostanoid metabolic process(GO:0006692) prostaglandin metabolic process(GO:0006693) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 2.0 | GO:0000377 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.9 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0051597 | response to methylmercury(GO:0051597) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.6 | 1.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.6 | 8.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.5 | 3.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.5 | 1.4 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.4 | 2.5 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.4 | 3.3 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.4 | 1.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.4 | 2.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.4 | 1.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.3 | 1.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.3 | 2.0 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 1.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.3 | 1.6 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.3 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.2 | 2.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.4 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.2 | 1.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.2 | 3.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 2.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 3.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.2 | 1.6 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.2 | 1.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.6 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.2 | 0.5 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.6 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.9 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.7 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 0.7 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.1 | 0.8 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 0.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 2.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 2.3 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.1 | 1.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 8.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 9.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.6 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.1 | 6.1 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 2.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 1.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.5 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.7 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.1 | 3.6 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 1.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.9 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.9 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 4.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.6 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 3.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 2.3 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.7 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.6 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 2.4 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0000784 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.2 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.8 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 2.6 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 29.5 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 2.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.4 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.0 | GO:0005774 | vacuolar membrane(GO:0005774) |
| 0.0 | 0.5 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.4 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 1.1 | GO:0030658 | transport vesicle membrane(GO:0030658) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.8 | 3.8 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.7 | 2.2 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.7 | 7.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.6 | 3.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.6 | 4.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.5 | 2.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.5 | 2.0 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.5 | 2.0 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.5 | 4.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.5 | 2.8 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.5 | 1.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.4 | 1.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.4 | 1.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.4 | 1.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 1.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 2.0 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.4 | 1.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.4 | 2.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.4 | 1.1 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.4 | 1.1 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.3 | 1.0 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.3 | 3.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 2.6 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.3 | 1.3 | GO:0031151 | histone methyltransferase activity (H3-K79 specific)(GO:0031151) |
| 0.3 | 2.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 3.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.3 | 1.2 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.3 | 2.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.3 | 1.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 1.9 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.3 | 1.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 0.8 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 4.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 7.0 | GO:0019003 | GDP binding(GO:0019003) |
| 0.2 | 0.6 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 1.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.2 | 1.5 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.2 | 2.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 1.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 1.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 1.9 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.3 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 1.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 4.0 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 1.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 0.7 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 3.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.4 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.1 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 3.0 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 1.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.0 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.1 | 0.4 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 1.7 | GO:0016229 | steroid dehydrogenase activity(GO:0016229) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.5 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 1.8 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.3 | GO:0030586 | [methionine synthase] reductase activity(GO:0030586) |
| 0.1 | 6.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 1.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.9 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.6 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 0.3 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.1 | 4.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 9.3 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.1 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.1 | 1.1 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 1.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 1.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 3.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 3.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 1.1 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 2.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 1.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 1.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 1.2 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.2 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 6.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0015126 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.6 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 1.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 1.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 9.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.1 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 8.2 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 2.3 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.9 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 3.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 2.8 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 1.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 2.4 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 2.1 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 3.4 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 4.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 7.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 8.0 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 5.8 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.2 | 1.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 1.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 6.8 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 5.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 0.6 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 3.0 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.7 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.5 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 3.5 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 1.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 5.4 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 4.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 1.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 1.6 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 2.2 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 1.4 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.1 | 1.3 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 1.2 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.5 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.4 | 5.0 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.3 | 3.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.3 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 1.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.2 | 3.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.2 | 1.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.2 | 1.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 1.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 1.9 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.6 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 2.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 6.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.4 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 2.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 2.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.1 | 1.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.9 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 3.3 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.1 | 1.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 2.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 0.6 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 1.7 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 0.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.1 | 3.4 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.1 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 1.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.8 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 2.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.7 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.6 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.7 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.1 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |