Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
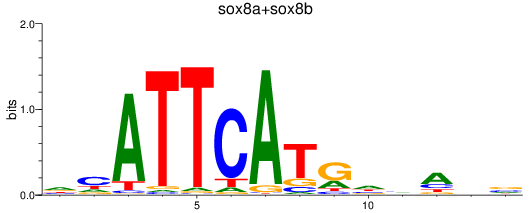
Results for sox8a+sox8b
Z-value: 0.26
Transcription factors associated with sox8a+sox8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox8b
|
ENSDARG00000037782 | SRY-box transcription factor 8b |
|
sox8a
|
ENSDARG00000105301 | SRY-box transcription factor 8a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox8a | dr11_v1_chr12_-_1085227_1085227 | -0.73 | 5.8e-04 | Click! |
| sox8b | dr11_v1_chr3_-_62403550_62403550 | -0.71 | 8.8e-04 | Click! |
Activity profile of sox8a+sox8b motif
Sorted Z-values of sox8a+sox8b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_27448606 | 0.57 |
ENSDART00000079251
|
vars2
|
valyl-tRNA synthetase 2, mitochondrial (putative) |
| chr12_-_11593436 | 0.55 |
ENSDART00000138954
|
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr12_-_33359052 | 0.53 |
ENSDART00000135943
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_-_33359654 | 0.53 |
ENSDART00000001907
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr22_-_817479 | 0.51 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr6_-_37743724 | 0.51 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr21_+_34088377 | 0.51 |
ENSDART00000170070
|
mtmr1b
|
myotubularin related protein 1b |
| chr21_+_34088110 | 0.50 |
ENSDART00000145123
ENSDART00000029599 ENSDART00000147519 |
mtmr1b
|
myotubularin related protein 1b |
| chr13_+_8892784 | 0.42 |
ENSDART00000075054
ENSDART00000143705 |
thada
|
thyroid adenoma associated |
| chr3_+_32663865 | 0.41 |
ENSDART00000190240
|
si:dkey-16l2.16
|
si:dkey-16l2.16 |
| chr23_-_27235403 | 0.41 |
ENSDART00000134418
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr19_+_11432330 | 0.41 |
ENSDART00000188065
|
PQLC1
|
PQ loop repeat containing 1 |
| chr16_+_46410520 | 0.39 |
ENSDART00000131072
|
rpz2
|
rapunzel 2 |
| chr19_+_27448625 | 0.38 |
ENSDART00000052352
|
ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr3_-_27647845 | 0.37 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr3_-_3413669 | 0.37 |
ENSDART00000113517
ENSDART00000179861 ENSDART00000115331 |
zgc:171446
|
zgc:171446 |
| chr10_+_5268054 | 0.37 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr9_-_50001606 | 0.37 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr2_+_15048410 | 0.36 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr20_-_39789036 | 0.34 |
ENSDART00000086405
ENSDART00000098253 |
rnf217
|
ring finger protein 217 |
| chr8_+_10862353 | 0.33 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr3_-_5067585 | 0.33 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr11_+_13176568 | 0.31 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr15_-_43327911 | 0.31 |
ENSDART00000077386
|
prss16
|
protease, serine, 16 (thymus) |
| chr5_+_31214341 | 0.30 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr18_+_45666489 | 0.30 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr14_+_38786298 | 0.30 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr2_+_24770435 | 0.30 |
ENSDART00000078854
|
mpv17l2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr11_-_43226255 | 0.29 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr10_+_7709724 | 0.28 |
ENSDART00000097670
|
ggcx
|
gamma-glutamyl carboxylase |
| chr22_+_9522971 | 0.27 |
ENSDART00000110048
|
strip1
|
striatin interacting protein 1 |
| chr3_+_39579393 | 0.26 |
ENSDART00000055170
|
cln3
|
ceroid-lipofuscinosis, neuronal 3 |
| chr11_+_30254556 | 0.25 |
ENSDART00000182316
|
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr10_+_15967643 | 0.25 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr7_+_25003851 | 0.25 |
ENSDART00000144526
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr16_-_21047483 | 0.25 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr7_+_17096281 | 0.24 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr8_-_7603516 | 0.23 |
ENSDART00000179826
ENSDART00000190153 |
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr7_-_50410524 | 0.22 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr8_-_7603700 | 0.20 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr12_+_34258139 | 0.20 |
ENSDART00000153127
|
socs3b
|
suppressor of cytokine signaling 3b |
| chr9_+_34952203 | 0.20 |
ENSDART00000121828
ENSDART00000142347 |
tfdp1a
|
transcription factor Dp-1, a |
| chr14_+_48862987 | 0.19 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr22_-_14262115 | 0.19 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr25_-_37248795 | 0.19 |
ENSDART00000087247
ENSDART00000154045 |
glg1a
|
golgi glycoprotein 1a |
| chr2_+_29925561 | 0.18 |
ENSDART00000163118
ENSDART00000183800 ENSDART00000180738 ENSDART00000139604 |
htr5aa
|
5-hydroxytryptamine (serotonin) receptor 5A, genome duplicate a |
| chr21_+_39673157 | 0.18 |
ENSDART00000100240
|
mrm3b
|
mitochondrial rRNA methyltransferase 3b |
| chr21_+_42717424 | 0.17 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr17_-_33707589 | 0.15 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr1_-_23308225 | 0.15 |
ENSDART00000137567
ENSDART00000008201 |
smim14
|
small integral membrane protein 14 |
| chr11_-_18015534 | 0.14 |
ENSDART00000181953
|
qrich1
|
glutamine-rich 1 |
| chr16_-_21047872 | 0.14 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr1_+_6646529 | 0.12 |
ENSDART00000144641
ENSDART00000103701 ENSDART00000138919 |
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr20_+_54383838 | 0.12 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr3_+_41726360 | 0.11 |
ENSDART00000154401
|
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr4_+_6833735 | 0.11 |
ENSDART00000136355
|
dock4b
|
dedicator of cytokinesis 4b |
| chr20_-_29505863 | 0.10 |
ENSDART00000148278
|
klf11a
|
Kruppel-like factor 11a |
| chr9_+_17423941 | 0.10 |
ENSDART00000112884
ENSDART00000155233 |
kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr20_+_25936370 | 0.09 |
ENSDART00000141265
|
znf106b
|
zinc finger protein 106b |
| chr2_+_23222939 | 0.09 |
ENSDART00000026800
|
kifap3b
|
kinesin-associated protein 3b |
| chr11_-_30138299 | 0.08 |
ENSDART00000172106
|
scml2
|
Scm polycomb group protein like 2 |
| chr20_-_3276339 | 0.08 |
ENSDART00000166831
|
rps6kc1
|
ribosomal protein S6 kinase polypeptide 1 |
| chr7_+_52753067 | 0.08 |
ENSDART00000053814
|
mtfmt
|
mitochondrial methionyl-tRNA formyltransferase |
| chr9_-_3934963 | 0.07 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr21_-_43328056 | 0.07 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr1_-_16688965 | 0.06 |
ENSDART00000079001
ENSDART00000178599 |
frg1
|
FSHD region gene 1 |
| chr6_-_48347724 | 0.06 |
ENSDART00000046973
|
capza1a
|
capping protein (actin filament) muscle Z-line, alpha 1a |
| chr15_+_3125136 | 0.06 |
ENSDART00000130968
|
rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr15_-_9419472 | 0.06 |
ENSDART00000122117
|
sacs
|
sacsin molecular chaperone |
| chr22_-_38621438 | 0.06 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr17_+_24687338 | 0.05 |
ENSDART00000135794
|
selenon
|
selenoprotein N |
| chr18_+_5917625 | 0.05 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr6_+_49926115 | 0.05 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr10_-_14545658 | 0.05 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr4_+_6833583 | 0.04 |
ENSDART00000165179
ENSDART00000186134 ENSDART00000174507 |
dock4b
|
dedicator of cytokinesis 4b |
| chr17_+_24604373 | 0.04 |
ENSDART00000082193
|
zmpste24
|
zinc metallopeptidase, STE24 homolog |
| chr9_-_43538328 | 0.04 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr4_+_40035116 | 0.04 |
ENSDART00000167808
|
BX649521.1
|
|
| chr4_+_39766931 | 0.03 |
ENSDART00000138663
|
si:dkeyp-85d8.1
|
si:dkeyp-85d8.1 |
| chr3_+_62327332 | 0.03 |
ENSDART00000158414
|
BX470259.1
|
|
| chr9_-_23147026 | 0.03 |
ENSDART00000167266
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr8_+_41238064 | 0.02 |
ENSDART00000111321
|
zgc:152830
|
zgc:152830 |
| chr8_-_14052349 | 0.02 |
ENSDART00000135811
|
atp2b3a
|
ATPase plasma membrane Ca2+ transporting 3a |
| chr9_+_23714406 | 0.02 |
ENSDART00000189445
|
gypc
|
glycophorin C (Gerbich blood group) |
| chr9_+_41156818 | 0.02 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr5_+_45139196 | 0.02 |
ENSDART00000113738
|
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_-_6567464 | 0.02 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr24_-_33873451 | 0.02 |
ENSDART00000159840
|
asic1c
|
acid-sensing (proton-gated) ion channel 1c |
| chr16_-_25607266 | 0.02 |
ENSDART00000192602
|
zgc:110410
|
zgc:110410 |
| chr17_-_39880725 | 0.02 |
ENSDART00000184030
|
zmp:0000000545
|
zmp:0000000545 |
| chr16_-_25606889 | 0.02 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr5_+_55265274 | 0.02 |
ENSDART00000131060
|
pcsk5a
|
proprotein convertase subtilisin/kexin type 5a |
| chr24_-_41220538 | 0.02 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr10_+_40629616 | 0.01 |
ENSDART00000147476
|
CR396590.9
|
|
| chr8_-_46386024 | 0.01 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr10_+_9595575 | 0.01 |
ENSDART00000091780
ENSDART00000184287 |
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr10_-_36214582 | 0.01 |
ENSDART00000166471
|
or109-11
|
odorant receptor, family D, subfamily 109, member 11 |
| chr5_+_41146786 | 0.01 |
ENSDART00000175766
|
sub1b
|
SUB1 homolog, transcriptional regulator b |
| chr6_+_7322587 | 0.01 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr19_+_12801940 | 0.01 |
ENSDART00000040073
|
mc5ra
|
melanocortin 5a receptor |
| chr14_-_25452503 | 0.01 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr10_-_7386475 | 0.00 |
ENSDART00000167963
|
nrg1
|
neuregulin 1 |
| chr10_-_43334914 | 0.00 |
ENSDART00000146660
ENSDART00000190288 |
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr14_+_31721171 | 0.00 |
ENSDART00000173034
|
adgrg4a
|
adhesion G protein-coupled receptor G4a |
| chr10_-_22249444 | 0.00 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr23_-_36418708 | 0.00 |
ENSDART00000132273
|
znf740b
|
zinc finger protein 740b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox8a+sox8b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.4 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.0 | 0.4 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.3 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.3 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.6 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.4 | GO:1990798 | pancreas regeneration(GO:1990798) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.4 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.0 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.0 | 1.1 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.3 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.1 | 0.2 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.1 | 0.6 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.6 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.2 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.3 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |