Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
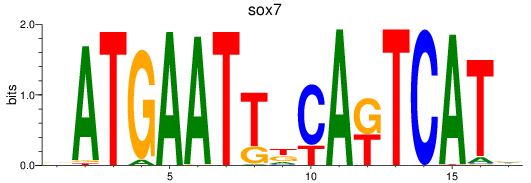
Results for sox7
Z-value: 0.29
Transcription factors associated with sox7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox7
|
ENSDARG00000030125 | SRY-box transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox7 | dr11_v1_chr20_+_19066858_19066858 | 0.62 | 5.7e-03 | Click! |
Activity profile of sox7 motif
Sorted Z-values of sox7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_44356707 | 0.71 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr11_+_44356504 | 0.69 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr20_-_4766645 | 0.69 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr24_+_35787629 | 0.66 |
ENSDART00000136721
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr2_-_24898226 | 0.64 |
ENSDART00000134936
|
si:dkey-149i17.9
|
si:dkey-149i17.9 |
| chr17_-_46817295 | 0.55 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr10_+_439692 | 0.53 |
ENSDART00000147740
|
zdhhc8a
|
zinc finger, DHHC-type containing 8a |
| chr7_-_52388734 | 0.50 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr14_+_6546516 | 0.50 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr17_-_46933567 | 0.49 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr11_-_41779281 | 0.42 |
ENSDART00000109204
|
megf6b
|
multiple EGF-like-domains 6b |
| chr15_-_21692630 | 0.38 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr22_-_29204960 | 0.38 |
ENSDART00000131386
|
pvalb7
|
parvalbumin 7 |
| chr6_-_16868834 | 0.37 |
ENSDART00000156503
|
pimr41
|
Pim proto-oncogene, serine/threonine kinase, related 41 |
| chr6_-_17266221 | 0.37 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr20_+_38276690 | 0.36 |
ENSDART00000061437
|
ccl38.6
|
chemokine (C-C motif) ligand 38, duplicate 6 |
| chr1_-_49649122 | 0.36 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr3_-_6709938 | 0.35 |
ENSDART00000172196
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr15_-_35252522 | 0.34 |
ENSDART00000144153
ENSDART00000059195 |
mff
|
mitochondrial fission factor |
| chr6_-_16735402 | 0.34 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr8_-_48741106 | 0.34 |
ENSDART00000164203
|
pimr180
|
Pim proto-oncogene, serine/threonine kinase, related 180 |
| chr16_+_10777116 | 0.33 |
ENSDART00000190902
|
atp1a3b
|
ATPase Na+/K+ transporting subunit alpha 3b |
| chr6_-_17052116 | 0.31 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr9_+_11587629 | 0.31 |
ENSDART00000102508
ENSDART00000146585 |
si:dkey-69o16.5
|
si:dkey-69o16.5 |
| chr14_+_8725216 | 0.29 |
ENSDART00000157630
|
pimr57
|
Pim proto-oncogene, serine/threonine kinase, related 57 |
| chr9_-_23033818 | 0.27 |
ENSDART00000022392
|
rnd3b
|
Rho family GTPase 3b |
| chr11_+_6010177 | 0.27 |
ENSDART00000170047
ENSDART00000022526 ENSDART00000161001 ENSDART00000188999 |
gtpbp3
|
GTP binding protein 3, mitochondrial |
| chr5_-_57169604 | 0.27 |
ENSDART00000136232
|
si:dkey-79c1.1
|
si:dkey-79c1.1 |
| chr9_+_32860345 | 0.26 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr7_+_18176162 | 0.26 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr15_-_23647078 | 0.26 |
ENSDART00000059366
|
ckmb
|
creatine kinase, muscle b |
| chr12_+_26621906 | 0.25 |
ENSDART00000158440
ENSDART00000046959 |
arhgap12b
|
Rho GTPase activating protein 12b |
| chr6_+_49081141 | 0.25 |
ENSDART00000131080
ENSDART00000050000 |
tshba
|
thyroid stimulating hormone, beta subunit, a |
| chr16_+_46111849 | 0.24 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr6_-_17179637 | 0.23 |
ENSDART00000153710
|
pimr19
|
Pim proto-oncogene, serine/threonine kinase, related 19 |
| chr22_+_29204885 | 0.23 |
ENSDART00000133409
ENSDART00000059820 |
ncf4
|
neutrophil cytosolic factor 4 |
| chr5_-_67911111 | 0.23 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr22_-_16658996 | 0.22 |
ENSDART00000138429
|
si:dkey-38n4.2
|
si:dkey-38n4.2 |
| chr21_+_19062124 | 0.22 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr25_-_17579701 | 0.21 |
ENSDART00000073684
|
mmp15a
|
matrix metallopeptidase 15a |
| chr6_-_16804001 | 0.21 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr22_-_29205327 | 0.21 |
ENSDART00000183161
ENSDART00000189515 |
pvalb7
|
parvalbumin 7 |
| chr4_+_47178124 | 0.21 |
ENSDART00000170816
|
si:dkey-26m3.2
|
si:dkey-26m3.2 |
| chr3_+_52737565 | 0.20 |
ENSDART00000108639
|
gmip
|
GEM interacting protein |
| chr5_+_34982318 | 0.19 |
ENSDART00000160504
|
ankra2
|
ankyrin repeat, family A (RFXANK-like), 2 |
| chr19_+_10860485 | 0.18 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
| chr20_+_50956369 | 0.16 |
ENSDART00000170854
|
gphnb
|
gephyrin b |
| chr6_-_16948040 | 0.15 |
ENSDART00000156433
|
pimr30
|
Pim proto-oncogene, serine/threonine kinase, related 30 |
| chr22_-_29191152 | 0.14 |
ENSDART00000132702
|
pvalb7
|
parvalbumin 7 |
| chr17_-_22010668 | 0.14 |
ENSDART00000031998
|
slc22a7b.1
|
solute carrier family 22 (organic anion transporter), member 7b, tandem duplicate 1 |
| chr9_+_32859967 | 0.14 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr4_-_77216726 | 0.13 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr1_+_36674584 | 0.13 |
ENSDART00000186772
ENSDART00000192274 |
ednraa
|
endothelin receptor type Aa |
| chr10_+_27068251 | 0.12 |
ENSDART00000012717
|
ehd1b
|
EH-domain containing 1b |
| chr16_+_27345383 | 0.11 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr18_-_18524624 | 0.11 |
ENSDART00000157834
|
lonp2
|
lon peptidase 2, peroxisomal |
| chr20_-_42534153 | 0.11 |
ENSDART00000061122
|
rfx6
|
regulatory factor X, 6 |
| chr5_+_65871688 | 0.09 |
ENSDART00000175217
|
FAM163B (1 of many)
|
family with sequence similarity 163 member B |
| chr16_+_40024883 | 0.09 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr14_+_45645024 | 0.09 |
ENSDART00000168278
|
si:ch211-276i12.4
|
si:ch211-276i12.4 |
| chr8_-_20291922 | 0.08 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr19_+_5480327 | 0.08 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr7_+_18878973 | 0.07 |
ENSDART00000037846
|
focad
|
focadhesin |
| chr13_+_835390 | 0.07 |
ENSDART00000171329
|
gsta.1
|
glutathione S-transferase, alpha tandem duplicate 1 |
| chr2_-_27385934 | 0.06 |
ENSDART00000139886
|
toe1
|
target of EGR1, exonuclease |
| chr4_+_33461796 | 0.06 |
ENSDART00000150445
|
si:dkey-247i3.1
|
si:dkey-247i3.1 |
| chr22_+_2239254 | 0.05 |
ENSDART00000131396
ENSDART00000135320 |
znf1144
|
zinc finger protein 1144 |
| chr22_-_26558166 | 0.05 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr2_-_45504798 | 0.05 |
ENSDART00000131412
|
gpsm2
|
G protein signaling modulator 2 |
| chr8_+_2787042 | 0.04 |
ENSDART00000155275
|
sh3glb2a
|
SH3-domain GRB2-like endophilin B2a |
| chr21_-_18932761 | 0.04 |
ENSDART00000140129
|
med15
|
mediator complex subunit 15 |
| chr10_-_23379146 | 0.04 |
ENSDART00000056369
|
cadm2a
|
cell adhesion molecule 2a |
| chr3_-_27065477 | 0.04 |
ENSDART00000185660
|
atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr2_-_24996441 | 0.04 |
ENSDART00000144795
|
slc35g2a
|
solute carrier family 35, member G2a |
| chr2_+_6991208 | 0.03 |
ENSDART00000182239
|
ddr2b
|
discoidin domain receptor tyrosine kinase 2b |
| chr20_+_10727022 | 0.03 |
ENSDART00000104185
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr4_-_71311438 | 0.02 |
ENSDART00000168021
|
si:dkeyp-123a12.2
|
si:dkeyp-123a12.2 |
| chr12_+_4114401 | 0.02 |
ENSDART00000025912
|
si:dkey-32n7.4
|
si:dkey-32n7.4 |
| chr21_+_20903244 | 0.02 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr12_-_4540564 | 0.02 |
ENSDART00000106566
|
CABZ01030107.1
|
|
| chr25_-_7999756 | 0.02 |
ENSDART00000159908
|
camk1db
|
calcium/calmodulin-dependent protein kinase 1Db |
| chr21_+_31811202 | 0.02 |
ENSDART00000189846
|
si:ch211-12m10.1
|
si:ch211-12m10.1 |
| chr12_+_30109698 | 0.02 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr16_-_31985965 | 0.01 |
ENSDART00000161944
|
si:dkey-40m6.14
|
si:dkey-40m6.14 |
| chr1_-_10669436 | 0.01 |
ENSDART00000152421
|
si:dkey-31e10.5
|
si:dkey-31e10.5 |
| chr20_+_37294112 | 0.01 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr7_-_35083184 | 0.01 |
ENSDART00000100253
ENSDART00000135250 ENSDART00000173511 |
agrp
|
agouti related neuropeptide |
| chr10_-_7913591 | 0.01 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr5_-_12824607 | 0.01 |
ENSDART00000190310
|
p2rx2
|
purinergic receptor P2X, ligand-gated ion channel, 2 |
| chr22_+_5682635 | 0.00 |
ENSDART00000140680
ENSDART00000131308 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr7_-_17412559 | 0.00 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 0.1 | 0.4 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.1 | 0.2 | GO:0072579 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.1 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.3 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 4.3 | GO:0046777 | protein autophosphorylation(GO:0046777) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045283 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.4 | GO:0016607 | nuclear speck(GO:0016607) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.2 | GO:0061599 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |