Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
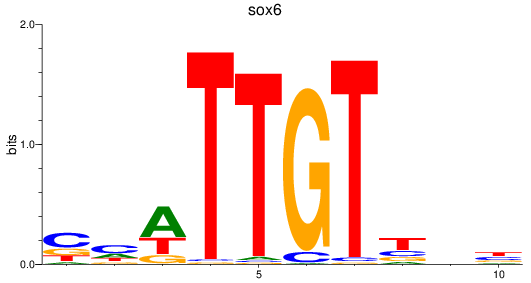
Results for sox6
Z-value: 1.22
Transcription factors associated with sox6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox6
|
ENSDARG00000015536 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox6 | dr11_v1_chr7_+_27250186_27250186 | -0.98 | 3.5e-12 | Click! |
Activity profile of sox6 motif
Sorted Z-values of sox6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2578026 | 6.46 |
ENSDART00000065821
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr12_+_48340133 | 4.90 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr17_-_2590222 | 4.56 |
ENSDART00000185711
|
CR759892.1
|
|
| chr1_-_33645967 | 3.94 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr22_-_10541372 | 3.83 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr12_-_33354409 | 3.79 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr19_-_27588842 | 3.56 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr17_-_2573021 | 3.45 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_24724233 | 3.38 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr10_+_15608326 | 3.08 |
ENSDART00000188770
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr24_-_19719240 | 2.76 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr16_-_54919260 | 2.52 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr10_+_2842923 | 2.41 |
ENSDART00000181895
|
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr14_-_41478265 | 2.22 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr2_+_26240631 | 2.21 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr2_+_32846602 | 2.21 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr13_+_33368503 | 2.18 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr15_+_12429206 | 2.08 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr22_-_10541712 | 2.07 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr13_+_46941930 | 1.99 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr1_-_354115 | 1.92 |
ENSDART00000141590
ENSDART00000098627 |
pros1
|
protein S |
| chr19_-_23249822 | 1.90 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr13_+_42309688 | 1.85 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr21_+_34119759 | 1.83 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr5_-_28149767 | 1.69 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr7_+_22801465 | 1.67 |
ENSDART00000052862
ENSDART00000173633 |
rbm4.1
|
RNA binding motif protein 4.1 |
| chr1_+_39995008 | 1.66 |
ENSDART00000166251
|
aip
|
aryl hydrocarbon receptor interacting protein |
| chr24_-_21343982 | 1.60 |
ENSDART00000012653
|
spice1
|
spindle and centriole associated protein 1 |
| chr9_+_21793565 | 1.56 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr13_-_33700461 | 1.56 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr22_-_37565348 | 1.51 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr3_+_49074008 | 1.47 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr3_-_25054002 | 1.41 |
ENSDART00000086768
|
ep300b
|
E1A binding protein p300 b |
| chr9_+_21795917 | 1.34 |
ENSDART00000169069
|
rev1
|
REV1, polymerase (DNA directed) |
| chr3_+_53156813 | 1.31 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr2_+_49457626 | 1.26 |
ENSDART00000129967
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr7_-_24520866 | 1.25 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr3_+_29641181 | 1.25 |
ENSDART00000151517
|
eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr19_-_27339844 | 1.25 |
ENSDART00000052358
ENSDART00000148238 ENSDART00000147661 ENSDART00000137346 |
znrd1
|
zinc ribbon domain containing 1 |
| chr8_-_49499457 | 1.24 |
ENSDART00000098326
|
opn7d
|
opsin 7, group member d |
| chr20_+_733510 | 1.21 |
ENSDART00000135066
ENSDART00000015558 ENSDART00000152782 |
myo6a
|
myosin VIa |
| chr22_+_4442473 | 1.20 |
ENSDART00000170751
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr10_+_39200213 | 1.18 |
ENSDART00000153727
|
ei24
|
etoposide induced 2.4 |
| chr25_+_34862225 | 1.17 |
ENSDART00000149782
|
CHST6
|
zgc:194879 |
| chr19_+_40069524 | 1.17 |
ENSDART00000151365
ENSDART00000140926 |
zmym4
|
zinc finger, MYM-type 4 |
| chr15_-_30853246 | 1.16 |
ENSDART00000112511
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr2_+_49457449 | 1.15 |
ENSDART00000185470
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr8_+_42941555 | 1.14 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr23_-_10175898 | 1.14 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr14_-_33978117 | 1.12 |
ENSDART00000128515
|
foxa
|
forkhead box A sequence |
| chr2_+_27386617 | 1.12 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr7_-_32895668 | 1.08 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr7_-_33868903 | 1.07 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr7_-_53117131 | 1.06 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr8_+_26879358 | 1.05 |
ENSDART00000132485
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr10_-_28028998 | 1.03 |
ENSDART00000023545
ENSDART00000143487 |
ints2
|
integrator complex subunit 2 |
| chr11_+_31324335 | 1.03 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr5_+_51833305 | 1.01 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr10_+_3428194 | 1.01 |
ENSDART00000081599
|
ptpn11a
|
protein tyrosine phosphatase, non-receptor type 11, a |
| chr17_+_24597001 | 1.00 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr3_+_26244353 | 1.00 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr15_+_29727799 | 0.97 |
ENSDART00000182006
|
zgc:153372
|
zgc:153372 |
| chr5_+_40835601 | 0.97 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr10_+_17371356 | 0.96 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr6_-_41138854 | 0.95 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr14_+_51056605 | 0.93 |
ENSDART00000159639
|
CABZ01078593.1
|
|
| chr19_-_43639331 | 0.93 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr15_-_9321921 | 0.91 |
ENSDART00000159676
ENSDART00000033411 |
trappc4
|
trafficking protein particle complex 4 |
| chr16_+_53387085 | 0.89 |
ENSDART00000154223
ENSDART00000101404 |
kif13a
|
kinesin family member 13A |
| chr25_+_33063762 | 0.87 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr16_+_24978203 | 0.86 |
ENSDART00000156579
|
si:dkeyp-84f3.5
|
si:dkeyp-84f3.5 |
| chr23_-_3759345 | 0.85 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr5_+_51833132 | 0.84 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr10_-_36633882 | 0.80 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr14_+_26247319 | 0.79 |
ENSDART00000192793
|
CCDC69
|
coiled-coil domain containing 69 |
| chr17_+_27158706 | 0.79 |
ENSDART00000151829
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr20_+_31217495 | 0.78 |
ENSDART00000020252
|
pdia6
|
protein disulfide isomerase family A, member 6 |
| chr10_+_29849497 | 0.77 |
ENSDART00000099994
ENSDART00000132212 |
hspa8
|
heat shock protein 8 |
| chr22_-_10440688 | 0.76 |
ENSDART00000111962
|
nol8
|
nucleolar protein 8 |
| chr17_+_5623514 | 0.75 |
ENSDART00000171220
ENSDART00000176083 |
CU571310.1
|
|
| chr5_+_55221593 | 0.75 |
ENSDART00000073638
|
tmc2a
|
transmembrane channel-like 2a |
| chr6_-_11362871 | 0.74 |
ENSDART00000151125
|
pcnt
|
pericentrin |
| chr25_-_22889519 | 0.73 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr10_+_5744941 | 0.72 |
ENSDART00000159769
ENSDART00000184734 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr21_+_36396864 | 0.68 |
ENSDART00000137309
|
gemin5
|
gem (nuclear organelle) associated protein 5 |
| chr19_+_7424347 | 0.68 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr22_+_4443689 | 0.65 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr25_+_28567381 | 0.64 |
ENSDART00000126552
|
SLC15A5
|
si:ch211-190o6.3 |
| chr19_+_16068445 | 0.62 |
ENSDART00000138709
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr7_+_24520518 | 0.62 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr13_+_15933168 | 0.62 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr6_-_19351495 | 0.61 |
ENSDART00000164287
|
grb2a
|
growth factor receptor-bound protein 2a |
| chr13_-_13294847 | 0.61 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr3_+_15773991 | 0.60 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr5_+_18012154 | 0.60 |
ENSDART00000139431
ENSDART00000048859 |
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr12_+_19866865 | 0.59 |
ENSDART00000045609
|
cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr8_+_16990120 | 0.59 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr19_+_24575077 | 0.58 |
ENSDART00000167469
|
si:dkeyp-92c9.4
|
si:dkeyp-92c9.4 |
| chr23_-_5685023 | 0.55 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr6_+_58280936 | 0.55 |
ENSDART00000155244
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr22_+_29113796 | 0.53 |
ENSDART00000150264
|
pla2g6
|
phospholipase A2, group VI (cytosolic, calcium-independent) |
| chr13_+_51579851 | 0.52 |
ENSDART00000163847
|
nkx6.2
|
NK6 homeobox 2 |
| chr9_-_32142311 | 0.50 |
ENSDART00000142768
|
ankrd44
|
ankyrin repeat domain 44 |
| chr8_+_8671229 | 0.50 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr7_-_49646251 | 0.48 |
ENSDART00000193674
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr9_-_18743012 | 0.47 |
ENSDART00000131626
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr2_-_30784198 | 0.46 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr9_+_8942258 | 0.43 |
ENSDART00000138836
|
ankrd10b
|
ankyrin repeat domain 10b |
| chr15_-_20779624 | 0.42 |
ENSDART00000181936
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr6_+_54888493 | 0.42 |
ENSDART00000113331
|
nav1b
|
neuron navigator 1b |
| chr11_+_34523132 | 0.40 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr25_+_13620555 | 0.39 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr11_+_14280598 | 0.39 |
ENSDART00000163033
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr22_-_18546241 | 0.38 |
ENSDART00000105404
ENSDART00000105405 |
cirbpb
|
cold inducible RNA binding protein b |
| chr5_+_35458346 | 0.37 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr9_-_18742704 | 0.37 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr6_-_25180438 | 0.36 |
ENSDART00000159696
|
lrrc8db
|
leucine rich repeat containing 8 VRAC subunit Db |
| chr6_-_52675630 | 0.36 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr7_+_24006875 | 0.35 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr12_-_7280551 | 0.34 |
ENSDART00000061633
|
zgc:171971
|
zgc:171971 |
| chr2_+_26479676 | 0.33 |
ENSDART00000056795
ENSDART00000144837 |
hectd3
|
HECT domain containing 3 |
| chr5_+_31480342 | 0.33 |
ENSDART00000098197
|
si:dkey-220k22.1
|
si:dkey-220k22.1 |
| chr1_-_20068155 | 0.32 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr7_-_73852594 | 0.32 |
ENSDART00000183194
|
zgc:165555
|
zgc:165555 |
| chr17_+_7534180 | 0.31 |
ENSDART00000187512
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr7_+_25103965 | 0.29 |
ENSDART00000192561
ENSDART00000173721 |
si:dkey-23i12.9
|
si:dkey-23i12.9 |
| chr1_-_31534089 | 0.29 |
ENSDART00000007770
|
lbx1b
|
ladybird homeobox 1b |
| chr9_-_22272181 | 0.29 |
ENSDART00000113174
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr9_-_22318511 | 0.28 |
ENSDART00000129295
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr1_-_18615063 | 0.28 |
ENSDART00000014916
|
klf3
|
Kruppel-like factor 3 (basic) |
| chr22_+_17531214 | 0.28 |
ENSDART00000161174
ENSDART00000088356 |
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr22_-_10570749 | 0.26 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr17_+_7534365 | 0.25 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr17_-_45386546 | 0.24 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr1_+_41690402 | 0.23 |
ENSDART00000177298
|
fbxo41
|
F-box protein 41 |
| chr6_-_7735153 | 0.23 |
ENSDART00000151545
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr9_-_9242777 | 0.23 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr8_+_17869225 | 0.23 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr12_+_40810418 | 0.23 |
ENSDART00000183393
|
CDH18
|
cadherin 18 |
| chr5_+_42912966 | 0.22 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr12_+_22657925 | 0.20 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr23_+_3538463 | 0.19 |
ENSDART00000172758
|
si:dkey-9l20.3
|
si:dkey-9l20.3 |
| chr12_+_19320657 | 0.17 |
ENSDART00000100075
ENSDART00000066389 |
tmem184ba
|
transmembrane protein 184ba |
| chr23_-_33944597 | 0.17 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr1_+_26480890 | 0.17 |
ENSDART00000164430
|
uso1
|
USO1 vesicle transport factor |
| chr25_+_13271458 | 0.16 |
ENSDART00000188307
|
AL772241.1
|
|
| chr21_+_349091 | 0.16 |
ENSDART00000183693
|
ZNF462
|
zinc finger protein 462 |
| chr4_+_65607540 | 0.16 |
ENSDART00000192218
|
si:dkey-205i10.2
|
si:dkey-205i10.2 |
| chr22_-_15693085 | 0.15 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr12_+_33151246 | 0.15 |
ENSDART00000162681
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr2_+_29257942 | 0.13 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr15_-_5563551 | 0.12 |
ENSDART00000099520
|
atg16l2
|
ATG16 autophagy related 16-like 2 (S. cerevisiae) |
| chr17_-_44776224 | 0.12 |
ENSDART00000156195
|
zdhhc22
|
zinc finger, DHHC-type containing 22 |
| chr21_-_9914745 | 0.12 |
ENSDART00000172124
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr21_-_41028665 | 0.11 |
ENSDART00000190531
|
plac8l1
|
PLAC8-like 1 |
| chr21_-_43550120 | 0.11 |
ENSDART00000151627
|
si:ch73-362m14.2
|
si:ch73-362m14.2 |
| chr4_-_5291256 | 0.10 |
ENSDART00000150864
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr9_+_50001746 | 0.10 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr24_-_29868151 | 0.09 |
ENSDART00000184802
|
aglb
|
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase b |
| chr11_+_10541258 | 0.08 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr10_-_25628555 | 0.08 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr3_-_9444749 | 0.08 |
ENSDART00000182191
|
FO904885.3
|
|
| chr17_-_45386823 | 0.07 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr20_-_40754794 | 0.07 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr7_+_31145386 | 0.07 |
ENSDART00000075407
ENSDART00000169462 |
fam189a1
|
family with sequence similarity 189, member A1 |
| chr18_+_47313899 | 0.07 |
ENSDART00000192389
ENSDART00000189592 ENSDART00000184281 |
barx2
|
BARX homeobox 2 |
| chr8_+_26205471 | 0.07 |
ENSDART00000131888
|
celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr3_+_3992590 | 0.07 |
ENSDART00000180211
ENSDART00000054840 |
BX005421.2
|
|
| chr10_-_42237304 | 0.06 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr5_+_19097194 | 0.06 |
ENSDART00000131918
|
unc13ba
|
unc-13 homolog Ba (C. elegans) |
| chr9_-_22339582 | 0.06 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr15_-_5239536 | 0.06 |
ENSDART00000081704
|
or128-2
|
odorant receptor, family E, subfamily 128, member 2 |
| chr11_-_7147540 | 0.06 |
ENSDART00000143942
|
bmp7a
|
bone morphogenetic protein 7a |
| chr15_+_31462664 | 0.05 |
ENSDART00000174267
ENSDART00000185665 ENSDART00000060087 |
or102-1
|
odorant receptor, family C, subfamily 102, member 1 |
| chr2_+_23731194 | 0.05 |
ENSDART00000155747
|
slc22a13a
|
solute carrier family 22 member 13a |
| chr5_+_25317061 | 0.05 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr12_-_30523216 | 0.05 |
ENSDART00000152896
ENSDART00000153191 |
plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr4_+_57046954 | 0.04 |
ENSDART00000171796
|
si:ch211-238e22.7
|
si:ch211-238e22.7 |
| chr13_-_11699530 | 0.04 |
ENSDART00000192161
|
slc39a8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr20_+_40237441 | 0.04 |
ENSDART00000168928
|
si:ch211-199i15.5
|
si:ch211-199i15.5 |
| chr9_+_3055566 | 0.04 |
ENSDART00000189906
ENSDART00000175891 ENSDART00000093021 |
ppp1r9ala
|
protein phosphatase 1 regulatory subunit 9A-like A |
| chr7_-_13362590 | 0.04 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr19_+_4051695 | 0.03 |
ENSDART00000166368
|
btr24
|
bloodthirsty-related gene family, member 24 |
| chr24_-_31194847 | 0.03 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr5_-_46505691 | 0.03 |
ENSDART00000111589
ENSDART00000122966 ENSDART00000166907 |
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr4_-_27301356 | 0.03 |
ENSDART00000100444
|
fam19a5a
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5a |
| chr5_-_33606729 | 0.02 |
ENSDART00000137073
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr25_+_26798673 | 0.01 |
ENSDART00000157235
|
ca12
|
carbonic anhydrase XII |
| chr16_-_42186093 | 0.01 |
ENSDART00000076030
|
fbl
|
fibrillarin |
| chr22_+_30631072 | 0.01 |
ENSDART00000059970
|
zmp:0000000606
|
zmp:0000000606 |
| chr15_-_41503392 | 0.01 |
ENSDART00000169351
|
si:dkey-230i18.2
|
si:dkey-230i18.2 |
| chr7_-_24373662 | 0.01 |
ENSDART00000173865
|
PTGR1 (1 of many)
|
si:dkey-11k2.7 |
| chr6_+_52889353 | 0.01 |
ENSDART00000174115
|
or137-9
|
odorant receptor, family H, subfamily 137, member 9 |
| chr4_+_29702703 | 0.01 |
ENSDART00000167771
|
si:ch211-214c20.1
|
si:ch211-214c20.1 |
| chr9_-_14084044 | 0.01 |
ENSDART00000141571
|
fer1l6
|
fer-1-like family member 6 |
| chr15_+_5287703 | 0.00 |
ENSDART00000157261
|
or122-1
|
odorant receptor, family E, subfamily 122, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.7 | 2.9 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.6 | 1.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.6 | 1.9 | GO:0032814 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.6 | 9.9 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.5 | 2.5 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.5 | 2.0 | GO:0051352 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.4 | 1.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.4 | 1.1 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.4 | 1.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 1.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.3 | 0.9 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.2 | 1.1 | GO:0090134 | adherens junction maintenance(GO:0034334) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.2 | 1.9 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 1.2 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.1 | 0.6 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 1.9 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 0.8 | GO:1902946 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.1 | 4.9 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.7 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.1 | 1.6 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.1 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.3 | GO:0002155 | regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.1 | 2.2 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.1 | 1.6 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.9 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.4 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.1 | 0.7 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.1 | 0.2 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 1.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.3 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 1.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.3 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 1.1 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.8 | GO:0030901 | midbrain development(GO:0030901) |
| 0.1 | 0.4 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 5.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.2 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.0 | 3.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 1.0 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.6 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 1.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0039703 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 2.2 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.9 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 2.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) wound healing, spreading of cells(GO:0044319) epiboly involved in wound healing(GO:0090505) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 1.4 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.1 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 1.7 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.7 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 1.1 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.7 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.5 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.3 | 1.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.3 | 2.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.3 | 0.8 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 1.0 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.2 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.1 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.9 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.0 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.9 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 2.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 3.9 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.7 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.9 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 8.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.6 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.9 | 9.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.3 | 1.0 | GO:0072591 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.3 | 1.0 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.3 | 3.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 2.2 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.3 | 0.9 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.2 | 0.7 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 2.9 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.0 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.3 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.9 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.8 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 1.2 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.1 | 0.8 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.8 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 1.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 2.1 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 1.4 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.2 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 1.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0004134 | glycogen debranching enzyme activity(GO:0004133) 4-alpha-glucanotransferase activity(GO:0004134) amylo-alpha-1,6-glucosidase activity(GO:0004135) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 3.5 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 1.3 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 1.1 | GO:0005254 | chloride channel activity(GO:0005254) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 5.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 0.9 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 1.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.4 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 3.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.9 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 2.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 0.8 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 0.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 2.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 1.1 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.5 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |