Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for sox5
Z-value: 0.96
Transcription factors associated with sox5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox5
|
ENSDARG00000011582 | SRY-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox5 | dr11_v1_chr4_-_17055782_17055782 | -0.54 | 2.0e-02 | Click! |
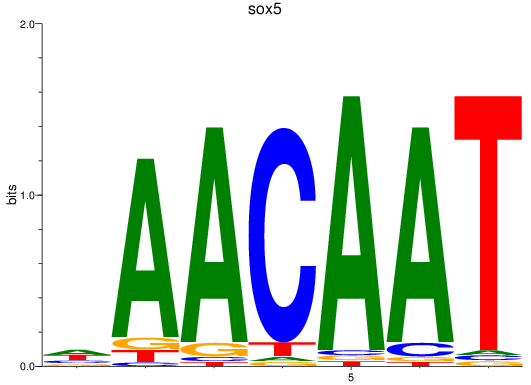
Activity profile of sox5 motif
Sorted Z-values of sox5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_868187 | 2.77 |
ENSDART00000186626
|
eomesa
|
eomesodermin homolog a |
| chr18_+_36066389 | 2.22 |
ENSDART00000059347
|
bckdha
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr19_+_10661520 | 1.98 |
ENSDART00000091813
ENSDART00000165653 |
ago3b
|
argonaute RISC catalytic component 3b |
| chr2_+_32846602 | 1.90 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr13_-_36844945 | 1.69 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr18_+_45571378 | 1.68 |
ENSDART00000077251
|
kifc3
|
kinesin family member C3 |
| chr19_+_41479990 | 1.60 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr17_+_28005763 | 1.59 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr9_-_14273652 | 1.57 |
ENSDART00000135458
|
abcb6b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6b |
| chr23_+_30730121 | 1.55 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr22_-_10541372 | 1.55 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr17_-_14966384 | 1.48 |
ENSDART00000105064
|
txndc16
|
thioredoxin domain containing 16 |
| chr5_+_40835601 | 1.46 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr21_+_13233377 | 1.46 |
ENSDART00000142569
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr12_+_48340133 | 1.45 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr21_+_7605803 | 1.42 |
ENSDART00000121813
|
wdr41
|
WD repeat domain 41 |
| chr21_+_13327527 | 1.38 |
ENSDART00000114294
|
snrpd3l
|
small nuclear ribonucleoprotein D3 polypeptide, like |
| chr12_-_33357655 | 1.37 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr15_-_41689981 | 1.37 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr6_-_42336987 | 1.31 |
ENSDART00000128777
ENSDART00000075601 |
fancd2
|
Fanconi anemia, complementation group D2 |
| chr19_+_7549854 | 1.29 |
ENSDART00000138866
ENSDART00000151758 |
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr3_-_25369557 | 1.28 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr11_-_25257045 | 1.27 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr24_-_19719240 | 1.26 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr14_-_2588216 | 1.25 |
ENSDART00000188082
ENSDART00000167293 |
fbxw11a
|
F-box and WD repeat domain containing 11a |
| chr1_+_45958904 | 1.24 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr12_-_17592215 | 1.24 |
ENSDART00000134597
|
usp42
|
ubiquitin specific peptidase 42 |
| chr11_-_34572202 | 1.24 |
ENSDART00000077883
|
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr11_-_25257595 | 1.23 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr16_-_39267185 | 1.21 |
ENSDART00000058550
ENSDART00000133642 |
gpd1l
|
glycerol-3-phosphate dehydrogenase 1 like |
| chr12_+_11650146 | 1.19 |
ENSDART00000150191
|
waplb
|
WAPL cohesin release factor b |
| chr5_+_24543862 | 1.19 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase H+ transporting V0 subunit a2b |
| chr2_+_16696052 | 1.19 |
ENSDART00000022356
ENSDART00000164329 |
ppp1r7
|
protein phosphatase 1, regulatory (inhibitor) subunit 7 |
| chr17_+_19626479 | 1.13 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr3_+_51684963 | 1.12 |
ENSDART00000091180
ENSDART00000183711 ENSDART00000159493 |
baiap2a
|
BAI1-associated protein 2a |
| chr18_-_12327426 | 1.12 |
ENSDART00000136992
ENSDART00000114024 |
fam107b
|
family with sequence similarity 107, member B |
| chr13_-_22961605 | 1.12 |
ENSDART00000143112
ENSDART00000057641 |
tspan15
|
tetraspanin 15 |
| chr15_-_41689684 | 1.12 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr15_+_24737599 | 1.12 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr2_+_44512324 | 1.11 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr11_+_26375979 | 1.10 |
ENSDART00000087652
ENSDART00000171748 ENSDART00000103513 ENSDART00000165931 ENSDART00000170043 |
cpne1
rbm12
|
copine I RNA binding motif protein 12 |
| chr7_-_60351537 | 1.09 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr23_+_24501918 | 1.08 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr15_-_35930070 | 1.07 |
ENSDART00000076229
|
irs1
|
insulin receptor substrate 1 |
| chr2_-_29923403 | 1.07 |
ENSDART00000144672
|
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr25_+_16895294 | 1.06 |
ENSDART00000159773
|
zgc:77158
|
zgc:77158 |
| chr14_+_49683767 | 1.06 |
ENSDART00000192515
|
tspan17
|
tetraspanin 17 |
| chr16_+_30117798 | 1.06 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr9_-_41040098 | 1.06 |
ENSDART00000008275
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr17_-_39185336 | 1.05 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr9_+_29520696 | 1.02 |
ENSDART00000144430
|
fdx1
|
ferredoxin 1 |
| chr9_+_42607138 | 1.01 |
ENSDART00000138133
ENSDART00000002027 |
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr25_-_29074064 | 1.00 |
ENSDART00000165603
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr15_+_29025090 | 1.00 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr23_+_19590006 | 1.00 |
ENSDART00000021231
|
slmapb
|
sarcolemma associated protein b |
| chr7_+_25003851 | 0.99 |
ENSDART00000144526
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr6_-_12900154 | 0.99 |
ENSDART00000080408
ENSDART00000150887 |
ical1
|
islet cell autoantigen 1-like |
| chr9_-_41040492 | 0.96 |
ENSDART00000163164
|
adat3
|
adenosine deaminase, tRNA-specific 3 |
| chr3_-_42086577 | 0.95 |
ENSDART00000083111
ENSDART00000187312 |
ttyh3a
|
tweety family member 3a |
| chr23_-_32157865 | 0.95 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr2_-_29923630 | 0.95 |
ENSDART00000158844
ENSDART00000031130 |
paxip1
|
PAX interacting (with transcription-activation domain) protein 1 |
| chr20_+_26943072 | 0.94 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr15_-_5799170 | 0.94 |
ENSDART00000142334
ENSDART00000171528 |
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr4_-_14624481 | 0.94 |
ENSDART00000137847
|
plxnb2a
|
plexin b2a |
| chr11_-_25461336 | 0.93 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr2_+_36620011 | 0.93 |
ENSDART00000177428
|
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr14_-_38889311 | 0.92 |
ENSDART00000186978
|
zgc:101583
|
zgc:101583 |
| chr8_+_13106760 | 0.92 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr2_-_21170517 | 0.92 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr14_-_41478265 | 0.91 |
ENSDART00000149886
ENSDART00000016002 |
tspan7
|
tetraspanin 7 |
| chr10_-_15879569 | 0.91 |
ENSDART00000136789
|
tjp2a
|
tight junction protein 2a (zona occludens 2) |
| chr19_-_46566430 | 0.90 |
ENSDART00000166668
|
ext1b
|
exostosin glycosyltransferase 1b |
| chr15_-_1036878 | 0.90 |
ENSDART00000123844
|
si:dkey-77f5.3
|
si:dkey-77f5.3 |
| chr6_+_32382743 | 0.89 |
ENSDART00000190009
|
dock7
|
dedicator of cytokinesis 7 |
| chr10_+_5954787 | 0.88 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr8_+_20488322 | 0.88 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr23_-_36446307 | 0.87 |
ENSDART00000136623
|
zgc:174906
|
zgc:174906 |
| chr13_+_25364324 | 0.86 |
ENSDART00000187471
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr2_+_4303125 | 0.86 |
ENSDART00000161211
ENSDART00000162269 |
cables1
|
Cdk5 and Abl enzyme substrate 1 |
| chr16_-_10223741 | 0.86 |
ENSDART00000188099
|
si:rp71-15i12.1
|
si:rp71-15i12.1 |
| chr18_+_19131773 | 0.84 |
ENSDART00000060766
|
rab11a
|
RAB11a, member RAS oncogene family |
| chr8_+_42941555 | 0.84 |
ENSDART00000183206
|
slc23a2
|
solute carrier family 23 (ascorbic acid transporter), member 2 |
| chr1_+_53321878 | 0.82 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr10_+_15255012 | 0.82 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr13_+_11440389 | 0.82 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr10_+_15255198 | 0.79 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr23_-_3759692 | 0.79 |
ENSDART00000028885
|
hmga1a
|
high mobility group AT-hook 1a |
| chr21_+_19547806 | 0.78 |
ENSDART00000159707
ENSDART00000184869 ENSDART00000181321 ENSDART00000058487 ENSDART00000058485 |
rai14
|
retinoic acid induced 14 |
| chr1_-_21714025 | 0.78 |
ENSDART00000129066
|
zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr20_+_54383838 | 0.77 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr3_+_26244353 | 0.77 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr11_+_2649891 | 0.76 |
ENSDART00000093052
|
si:ch211-160o17.4
|
si:ch211-160o17.4 |
| chr19_+_29808699 | 0.76 |
ENSDART00000051799
ENSDART00000164205 |
hdac1
|
histone deacetylase 1 |
| chr12_+_33403694 | 0.76 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr13_+_25364753 | 0.76 |
ENSDART00000027428
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr15_+_29662401 | 0.75 |
ENSDART00000135540
|
nrip1a
|
nuclear receptor interacting protein 1a |
| chr23_-_36003282 | 0.75 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr7_+_24528866 | 0.75 |
ENSDART00000180552
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr14_-_38889840 | 0.75 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr13_-_32898962 | 0.74 |
ENSDART00000163757
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr10_-_14929630 | 0.74 |
ENSDART00000121892
ENSDART00000044756 ENSDART00000128579 ENSDART00000147653 |
smad2
|
SMAD family member 2 |
| chr23_-_3759345 | 0.74 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr2_+_22363824 | 0.74 |
ENSDART00000163172
|
hs2st1a
|
heparan sulfate 2-O-sulfotransferase 1a |
| chr13_-_25196758 | 0.73 |
ENSDART00000184722
|
adka
|
adenosine kinase a |
| chr20_-_31238313 | 0.73 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr25_+_26441054 | 0.72 |
ENSDART00000158607
|
dapk2b
|
death-associated protein kinase 2b |
| chr19_+_20793388 | 0.72 |
ENSDART00000142463
|
txnl4a
|
thioredoxin-like 4A |
| chr6_+_18520859 | 0.72 |
ENSDART00000158263
|
si:dkey-10p5.10
|
si:dkey-10p5.10 |
| chr19_+_29808471 | 0.71 |
ENSDART00000186428
|
hdac1
|
histone deacetylase 1 |
| chr16_+_27614989 | 0.71 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr3_+_54047342 | 0.71 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr17_+_16192486 | 0.71 |
ENSDART00000156832
|
kif13ba
|
kinesin family member 13Ba |
| chr5_+_25762271 | 0.71 |
ENSDART00000181323
|
tmem2
|
transmembrane protein 2 |
| chr2_+_2967255 | 0.70 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr16_-_21047483 | 0.70 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr12_-_23365737 | 0.69 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr14_-_6402769 | 0.69 |
ENSDART00000121552
|
slc44a1b
|
solute carrier family 44 (choline transporter), member 1b |
| chr10_-_36625752 | 0.68 |
ENSDART00000138344
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr6_-_46742455 | 0.67 |
ENSDART00000011970
|
zgc:66479
|
zgc:66479 |
| chr17_+_28706946 | 0.66 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr24_+_32411753 | 0.66 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr25_+_28253844 | 0.66 |
ENSDART00000151891
|
cadps2
|
Ca++-dependent secretion activator 2 |
| chr2_+_35880600 | 0.65 |
ENSDART00000004277
|
lamc1
|
laminin, gamma 1 |
| chr8_-_49766205 | 0.65 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr19_+_13410903 | 0.65 |
ENSDART00000165033
ENSDART00000168672 |
srsf4
|
serine/arginine-rich splicing factor 4 |
| chr19_-_5058515 | 0.65 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr3_-_32873641 | 0.64 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr17_+_21486047 | 0.62 |
ENSDART00000104608
|
pla2g4f.2
|
phospholipase A2, group IVF, tandem duplicate 2 |
| chr15_-_21669618 | 0.62 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr8_-_14604606 | 0.62 |
ENSDART00000090254
ENSDART00000188953 |
cep350
|
centrosomal protein 350 |
| chr25_+_388258 | 0.62 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr21_+_19368720 | 0.62 |
ENSDART00000187759
ENSDART00000185829 ENSDART00000158471 ENSDART00000168728 |
btc
|
betacellulin, epidermal growth factor family member |
| chr3_-_37148594 | 0.60 |
ENSDART00000140855
|
mlx
|
MLX, MAX dimerization protein |
| chr16_-_21047872 | 0.60 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr3_+_40407352 | 0.59 |
ENSDART00000155112
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr18_-_20466061 | 0.59 |
ENSDART00000060311
|
paqr5a
|
progestin and adipoQ receptor family member Va |
| chr2_+_31308587 | 0.58 |
ENSDART00000027090
|
clul1
|
clusterin-like 1 (retinal) |
| chr10_+_8847033 | 0.58 |
ENSDART00000140894
|
itga2.1
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 1 |
| chr5_-_20893993 | 0.58 |
ENSDART00000137603
|
si:ch211-225b11.4
|
si:ch211-225b11.4 |
| chr5_-_38342992 | 0.58 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr3_+_31058464 | 0.57 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr12_+_27704015 | 0.57 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr10_+_31244619 | 0.57 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr6_-_6258451 | 0.56 |
ENSDART00000081966
ENSDART00000125918 |
rtn4a
|
reticulon 4a |
| chr20_+_21391181 | 0.56 |
ENSDART00000185158
ENSDART00000049586 ENSDART00000024922 |
jag2b
|
jagged 2b |
| chr17_+_24597001 | 0.56 |
ENSDART00000191834
|
rlf
|
rearranged L-myc fusion |
| chr3_-_30186296 | 0.54 |
ENSDART00000134395
ENSDART00000077057 ENSDART00000017422 |
tbc1d17
|
TBC1 domain family, member 17 |
| chr15_+_29728377 | 0.54 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr21_-_42876565 | 0.54 |
ENSDART00000126480
|
zmp:0000001268
|
zmp:0000001268 |
| chr3_-_26244256 | 0.52 |
ENSDART00000103741
|
ppp4ca
|
protein phosphatase 4, catalytic subunit a |
| chr17_-_24889837 | 0.52 |
ENSDART00000187980
|
gale
|
UDP-galactose-4-epimerase |
| chr6_-_51771634 | 0.50 |
ENSDART00000073847
|
blcap
|
bladder cancer associated protein |
| chr23_-_31428763 | 0.50 |
ENSDART00000053545
|
zgc:153284
|
zgc:153284 |
| chr3_+_19685873 | 0.50 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr23_+_43770149 | 0.49 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr5_-_24543526 | 0.48 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr22_-_29906764 | 0.47 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr7_+_67749251 | 0.47 |
ENSDART00000167562
|
dhx38
|
DEAH (Asp-Glu-Ala-His) box polypeptide 38 |
| chr6_-_45869127 | 0.47 |
ENSDART00000062459
ENSDART00000180563 |
rbm19
|
RNA binding motif protein 19 |
| chr8_+_17869225 | 0.47 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr16_+_35535375 | 0.47 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr22_+_10232527 | 0.46 |
ENSDART00000139297
|
si:dkeyp-87e7.4
|
si:dkeyp-87e7.4 |
| chr23_+_20640875 | 0.45 |
ENSDART00000147382
|
uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr18_+_30421528 | 0.45 |
ENSDART00000140908
|
gse1
|
Gse1 coiled-coil protein |
| chr9_-_43213229 | 0.45 |
ENSDART00000139775
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr6_-_10728057 | 0.44 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr10_-_8672820 | 0.44 |
ENSDART00000080763
|
si:dkey-27b3.2
|
si:dkey-27b3.2 |
| chr10_+_9222476 | 0.44 |
ENSDART00000064973
ENSDART00000145879 |
paqr3b
|
progestin and adipoQ receptor family member IIIb |
| chr4_-_2059233 | 0.44 |
ENSDART00000188177
ENSDART00000129521 ENSDART00000082289 |
cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr22_-_20342260 | 0.44 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr23_+_22819971 | 0.44 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr16_+_40954481 | 0.44 |
ENSDART00000058587
|
gbp
|
glycogen synthase kinase binding protein |
| chr17_+_31621788 | 0.43 |
ENSDART00000111629
ENSDART00000157148 |
cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr21_+_13244450 | 0.43 |
ENSDART00000146062
|
specc1lb
|
sperm antigen with calponin homology and coiled-coil domains 1-like b |
| chr17_-_35278763 | 0.43 |
ENSDART00000063437
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr23_-_5685023 | 0.42 |
ENSDART00000148680
ENSDART00000149365 |
tnnt2a
|
troponin T type 2a (cardiac) |
| chr19_+_37848830 | 0.41 |
ENSDART00000042276
ENSDART00000180872 |
nxph1
|
neurexophilin 1 |
| chr23_-_17003533 | 0.41 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr17_+_49281597 | 0.41 |
ENSDART00000155599
|
zgc:113176
|
zgc:113176 |
| chr16_-_36196700 | 0.40 |
ENSDART00000172324
|
capn7
|
calpain 7 |
| chr18_-_12451772 | 0.40 |
ENSDART00000175083
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr8_-_43677762 | 0.39 |
ENSDART00000167762
|
ep400
|
E1A binding protein p400 |
| chr5_+_64397454 | 0.39 |
ENSDART00000015940
|
edf1
|
endothelial differentiation-related factor 1 |
| chr23_-_15916316 | 0.39 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr2_+_24867534 | 0.37 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr7_-_32020858 | 0.37 |
ENSDART00000023568
|
kif18a
|
kinesin family member 18A |
| chr2_+_35806672 | 0.37 |
ENSDART00000137384
|
rasal2
|
RAS protein activator like 2 |
| chr25_+_7321675 | 0.36 |
ENSDART00000104712
ENSDART00000142934 |
hmg20a
|
high mobility group 20A |
| chr7_-_33351485 | 0.36 |
ENSDART00000146420
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr23_+_17102960 | 0.36 |
ENSDART00000053414
|
commd7
|
COMM domain containing 7 |
| chr14_-_44773864 | 0.36 |
ENSDART00000158386
|
si:dkey-109l4.3
|
si:dkey-109l4.3 |
| chr6_+_46406565 | 0.35 |
ENSDART00000168440
ENSDART00000131203 ENSDART00000138567 ENSDART00000132845 |
pbrm1l
|
polybromo 1, like |
| chr8_-_39978767 | 0.35 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr7_+_38090515 | 0.34 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr19_-_38872650 | 0.34 |
ENSDART00000146641
|
adgrb2
|
adhesion G protein-coupled receptor B2 |
| chr19_-_6988837 | 0.34 |
ENSDART00000145741
ENSDART00000167640 |
znf384l
|
zinc finger protein 384 like |
| chr13_+_15933168 | 0.34 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr4_+_23134366 | 0.33 |
ENSDART00000185788
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr5_-_18962794 | 0.33 |
ENSDART00000145210
|
ankle2
|
ankyrin repeat and LEM domain containing 2 |
| chr23_+_25201077 | 0.33 |
ENSDART00000136675
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.8 | GO:0055109 | invagination involved in gastrulation with mouth forming second(GO:0055109) morphogenesis of an epithelial fold(GO:0060571) |
| 0.6 | 1.9 | GO:0097095 | frontonasal suture morphogenesis(GO:0097095) |
| 0.4 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.4 | 1.2 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.3 | 1.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.3 | 1.6 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.3 | 1.6 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 1.2 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.3 | 0.8 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.3 | 0.8 | GO:0015882 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.3 | 1.3 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 0.7 | GO:0060031 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) mediolateral intercalation(GO:0060031) |
| 0.2 | 1.1 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.2 | 1.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.2 | 1.5 | GO:0033687 | osteoblast proliferation(GO:0033687) |
| 0.2 | 2.0 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.2 | 0.6 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 1.3 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.6 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.1 | 2.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.4 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 1.1 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.1 | 1.0 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.1 | 0.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 1.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 0.6 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.1 | 1.0 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.1 | 0.5 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 1.7 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.2 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 1.0 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 0.6 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 0.4 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 1.1 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.1 | 0.3 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.1 | 0.8 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.5 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 1.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.9 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.4 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.1 | 0.5 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.1 | 0.7 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.8 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.2 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.8 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.6 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) anterior lateral line nerve development(GO:0048909) |
| 0.0 | 1.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.2 | GO:0071387 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.0 | 1.1 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 1.0 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.5 | GO:0002031 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) G-protein coupled receptor internalization(GO:0002031) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0051099 | positive regulation of DNA binding(GO:0043388) positive regulation of binding(GO:0051099) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.9 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.3 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.0 | 1.3 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.3 | GO:0051928 | positive regulation of calcium ion transport(GO:0051928) |
| 0.0 | 0.5 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 1.7 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.7 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 1.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 2.1 | GO:1990778 | protein localization to plasma membrane(GO:0072659) protein localization to cell periphery(GO:1990778) |
| 0.0 | 0.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.6 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.5 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 1.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.3 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 1.2 | GO:0006909 | phagocytosis(GO:0006909) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.4 | 3.6 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.3 | 1.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 0.7 | GO:0031213 | RSF complex(GO:0031213) |
| 0.2 | 1.6 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.2 | 1.4 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.4 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 1.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 1.6 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.1 | 0.4 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.1 | 1.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 1.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.5 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.3 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.3 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 2.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.9 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0034707 | chloride channel complex(GO:0034707) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.7 | 2.0 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.4 | 1.6 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.4 | 1.6 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.4 | 1.6 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.4 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.3 | 1.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 1.6 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 1.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.3 | 0.8 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 2.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.2 | 1.0 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.2 | 0.7 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.2 | 0.9 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.2 | 1.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.5 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.2 | 2.2 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 0.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 1.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 1.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.7 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 1.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 1.0 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 1.2 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 1.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.5 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 1.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.1 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.3 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.6 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.3 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.3 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.5 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 0.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.6 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.1 | 1.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 1.8 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.8 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.8 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 1.6 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 3.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.2 | 2.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 1.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 1.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.1 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.1 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.7 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |