Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
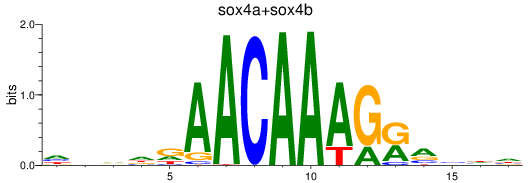
Results for sox4a+sox4b
Z-value: 0.57
Transcription factors associated with sox4a+sox4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox4a
|
ENSDARG00000004588 | SRY-box transcription factor 4a |
|
sox4b
|
ENSDARG00000098834 | SRY-box transcription factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox4b | dr11_v1_chr16_+_68069_68124 | -0.72 | 7.1e-04 | Click! |
| sox4a | dr11_v1_chr19_-_28789404_28789409 | -0.46 | 5.5e-02 | Click! |
Activity profile of sox4a+sox4b motif
Sorted Z-values of sox4a+sox4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_18152407 | 0.91 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr24_-_37568359 | 0.68 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr22_+_8979955 | 0.62 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr13_+_22675802 | 0.59 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr8_-_49908978 | 0.58 |
ENSDART00000172642
|
agtpbp1
|
ATP/GTP binding protein 1 |
| chr1_+_44173245 | 0.52 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr16_-_6944927 | 0.47 |
ENSDART00000149620
|
pmvk
|
phosphomevalonate kinase |
| chr12_-_33646010 | 0.47 |
ENSDART00000111259
|
tmem94
|
transmembrane protein 94 |
| chr20_+_34770197 | 0.46 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr13_+_39188737 | 0.46 |
ENSDART00000083641
|
fam135a
|
family with sequence similarity 135, member A |
| chr19_-_46037835 | 0.46 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr5_+_44846434 | 0.46 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr11_+_807153 | 0.45 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr15_+_22435460 | 0.44 |
ENSDART00000031976
|
tmem136a
|
transmembrane protein 136a |
| chr7_+_16348835 | 0.44 |
ENSDART00000002449
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr19_+_7424347 | 0.43 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr6_-_54126463 | 0.43 |
ENSDART00000161059
|
tusc2a
|
tumor suppressor candidate 2a |
| chr10_-_35103208 | 0.42 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr15_+_12429206 | 0.40 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr24_-_35707552 | 0.40 |
ENSDART00000165199
|
mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr14_+_6963312 | 0.40 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr6_+_12968101 | 0.39 |
ENSDART00000013781
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr14_+_6962271 | 0.39 |
ENSDART00000148447
ENSDART00000149114 ENSDART00000149492 ENSDART00000148394 |
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr7_-_24995631 | 0.38 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr16_+_6944564 | 0.37 |
ENSDART00000104252
|
eaf1
|
ELL associated factor 1 |
| chr23_-_10745288 | 0.37 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr2_-_24069331 | 0.37 |
ENSDART00000156972
ENSDART00000181691 ENSDART00000157041 |
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr6_-_8244474 | 0.36 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr16_-_33105677 | 0.36 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr11_+_11152214 | 0.36 |
ENSDART00000148030
|
ly75
|
lymphocyte antigen 75 |
| chr9_+_12948511 | 0.35 |
ENSDART00000135797
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr12_-_5188413 | 0.34 |
ENSDART00000161988
|
fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr5_+_44846280 | 0.34 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr2_+_2967255 | 0.34 |
ENSDART00000167649
ENSDART00000166449 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr17_-_2036850 | 0.33 |
ENSDART00000186048
ENSDART00000188838 ENSDART00000186423 |
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr8_+_17869225 | 0.33 |
ENSDART00000080079
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr3_-_13955878 | 0.33 |
ENSDART00000166804
|
gcdhb
|
glutaryl-CoA dehydrogenase b |
| chr3_-_25054002 | 0.32 |
ENSDART00000086768
|
ep300b
|
E1A binding protein p300 b |
| chr25_-_12902242 | 0.31 |
ENSDART00000164733
|
sept15
|
septin 15 |
| chr12_+_17436904 | 0.31 |
ENSDART00000079130
|
atad1b
|
ATPase family, AAA domain containing 1b |
| chr8_+_29636431 | 0.31 |
ENSDART00000133047
|
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr6_+_7322587 | 0.31 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr19_+_7636941 | 0.30 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr14_-_44773864 | 0.30 |
ENSDART00000158386
|
si:dkey-109l4.3
|
si:dkey-109l4.3 |
| chr14_+_33525196 | 0.30 |
ENSDART00000085335
|
zdhhc9
|
zinc finger, DHHC-type containing 9 |
| chr11_-_2250767 | 0.30 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr13_-_21650404 | 0.30 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr9_-_36924388 | 0.30 |
ENSDART00000059756
|
ralba
|
v-ral simian leukemia viral oncogene homolog Ba (ras related) |
| chr2_-_23538794 | 0.29 |
ENSDART00000143857
|
si:dkey-58b18.10
|
si:dkey-58b18.10 |
| chr16_+_6944311 | 0.28 |
ENSDART00000144763
|
eaf1
|
ELL associated factor 1 |
| chr22_-_8006342 | 0.28 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr2_-_24898226 | 0.28 |
ENSDART00000134936
|
si:dkey-149i17.9
|
si:dkey-149i17.9 |
| chr2_-_23600821 | 0.28 |
ENSDART00000146217
|
BX677666.1
|
|
| chr20_+_6533260 | 0.28 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr24_-_31194847 | 0.28 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr16_-_20932896 | 0.27 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr4_+_9177997 | 0.27 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr12_+_19320657 | 0.27 |
ENSDART00000100075
ENSDART00000066389 |
tmem184ba
|
transmembrane protein 184ba |
| chr18_+_8231138 | 0.27 |
ENSDART00000140193
|
arsa
|
arylsulfatase A |
| chr18_+_39327010 | 0.27 |
ENSDART00000012164
|
tmod2
|
tropomodulin 2 |
| chr16_-_54919260 | 0.26 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr17_+_50074372 | 0.26 |
ENSDART00000113644
|
vps39
|
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr10_-_1697037 | 0.26 |
ENSDART00000125188
ENSDART00000002985 |
srsf9
|
serine/arginine-rich splicing factor 9 |
| chr8_+_4803906 | 0.25 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr22_-_22301672 | 0.25 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr23_+_35714574 | 0.25 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr21_+_17110598 | 0.25 |
ENSDART00000101282
ENSDART00000191864 |
bcr
|
breakpoint cluster region |
| chr2_+_47754419 | 0.25 |
ENSDART00000188221
|
mbnl1
|
muscleblind-like splicing regulator 1 |
| chr24_-_12983829 | 0.25 |
ENSDART00000133324
|
dcaf11
|
ddb1 and cul4 associated factor 11 |
| chr13_-_27660955 | 0.25 |
ENSDART00000188651
ENSDART00000134494 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr17_+_7534365 | 0.24 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr25_-_27665978 | 0.24 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr1_-_44701313 | 0.24 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr12_+_13348918 | 0.24 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr17_-_45378473 | 0.24 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr9_+_48007081 | 0.23 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr12_-_20616160 | 0.23 |
ENSDART00000105362
|
snx11
|
sorting nexin 11 |
| chr5_-_28149767 | 0.23 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr25_+_12494079 | 0.23 |
ENSDART00000163508
|
map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr25_+_26895394 | 0.23 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr6_+_7249531 | 0.23 |
ENSDART00000125912
ENSDART00000083424 ENSDART00000049695 ENSDART00000136088 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr1_+_44173506 | 0.22 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr23_-_36306337 | 0.22 |
ENSDART00000142760
ENSDART00000136929 ENSDART00000143340 |
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr2_+_18988407 | 0.22 |
ENSDART00000170216
|
glula
|
glutamate-ammonia ligase (glutamine synthase) a |
| chr10_-_2788668 | 0.22 |
ENSDART00000131749
ENSDART00000124356 ENSDART00000085031 |
ash2l
|
ash2 (absent, small, or homeotic)-like (Drosophila) |
| chr13_+_48358467 | 0.21 |
ENSDART00000171080
ENSDART00000162531 |
msh6
|
mutS homolog 6 (E. coli) |
| chr7_-_24149670 | 0.21 |
ENSDART00000005884
|
mmp14a
|
matrix metallopeptidase 14a (membrane-inserted) |
| chr7_-_26308098 | 0.21 |
ENSDART00000146440
ENSDART00000146935 ENSDART00000164627 |
zgc:77439
|
zgc:77439 |
| chr21_-_9914745 | 0.21 |
ENSDART00000172124
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr11_-_38492564 | 0.20 |
ENSDART00000102850
|
elk4
|
ELK4, ETS-domain protein |
| chr6_-_9282080 | 0.20 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr24_+_17142881 | 0.20 |
ENSDART00000177272
ENSDART00000192259 ENSDART00000191029 |
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr16_+_6750756 | 0.20 |
ENSDART00000149720
|
znf236
|
zinc finger protein 236 |
| chr7_-_32629458 | 0.20 |
ENSDART00000001376
|
arl14ep
|
ADP-ribosylation factor-like 14 effector protein |
| chr4_-_20135406 | 0.20 |
ENSDART00000161343
|
cep83
|
centrosomal protein 83 |
| chr10_-_25628555 | 0.20 |
ENSDART00000143978
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr8_-_20862443 | 0.19 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr3_-_36364903 | 0.19 |
ENSDART00000028883
|
gna13b
|
guanine nucleotide binding protein (G protein), alpha 13b |
| chr17_+_25849332 | 0.19 |
ENSDART00000191994
|
acss1
|
acyl-CoA synthetase short chain family member 1 |
| chr9_+_54179306 | 0.19 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr16_-_24612871 | 0.19 |
ENSDART00000155614
ENSDART00000154787 ENSDART00000155983 ENSDART00000156519 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr17_+_7534180 | 0.19 |
ENSDART00000187512
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr4_+_9178913 | 0.18 |
ENSDART00000168558
|
nfyba
|
nuclear transcription factor Y, beta a |
| chr16_+_29586468 | 0.18 |
ENSDART00000148926
|
mcl1b
|
MCL1, BCL2 family apoptosis regulator b |
| chr2_-_7696287 | 0.18 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr24_+_24923166 | 0.18 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr16_+_19013257 | 0.18 |
ENSDART00000144102
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr3_-_16110100 | 0.18 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr14_+_39258569 | 0.18 |
ENSDART00000103298
|
diaph2
|
diaphanous-related formin 2 |
| chr22_-_22337382 | 0.18 |
ENSDART00000144684
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr22_-_33916620 | 0.18 |
ENSDART00000191276
|
CR974456.1
|
|
| chr11_-_38492269 | 0.17 |
ENSDART00000065613
|
elk4
|
ELK4, ETS-domain protein |
| chr14_-_28430505 | 0.17 |
ENSDART00000192373
|
nek12
|
NIMA-related kinase 12 |
| chr9_-_54840124 | 0.17 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr14_-_5642371 | 0.17 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr19_-_17774875 | 0.17 |
ENSDART00000151133
ENSDART00000130695 |
top2b
|
DNA topoisomerase II beta |
| chr15_+_34963316 | 0.17 |
ENSDART00000153840
|
si:ch73-95l15.5
|
si:ch73-95l15.5 |
| chr19_-_7540821 | 0.17 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr4_-_20511595 | 0.16 |
ENSDART00000185806
|
rassf8b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8b |
| chr3_-_16110351 | 0.16 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr19_-_30800004 | 0.16 |
ENSDART00000128560
ENSDART00000045504 ENSDART00000125893 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr1_+_17376922 | 0.16 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr25_+_3507368 | 0.16 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr25_+_25737386 | 0.16 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr23_+_12361899 | 0.16 |
ENSDART00000143728
|
pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr17_-_43012390 | 0.16 |
ENSDART00000155615
|
atg2b
|
autophagy related 2B |
| chr15_-_15469079 | 0.15 |
ENSDART00000132637
ENSDART00000004220 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr7_-_24875421 | 0.15 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr22_-_7025393 | 0.15 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr5_+_1965296 | 0.15 |
ENSDART00000156224
|
dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr24_+_9696760 | 0.15 |
ENSDART00000140200
ENSDART00000187411 |
topbp1
|
DNA topoisomerase II binding protein 1 |
| chr9_+_33334501 | 0.15 |
ENSDART00000006867
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr12_-_22400999 | 0.15 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr5_+_15203421 | 0.15 |
ENSDART00000040826
|
tbx1
|
T-box 1 |
| chr14_-_31151148 | 0.14 |
ENSDART00000026569
|
gpc4
|
glypican 4 |
| chr7_+_10563017 | 0.14 |
ENSDART00000193520
ENSDART00000173125 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr23_+_36306539 | 0.14 |
ENSDART00000053267
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr10_-_25591194 | 0.13 |
ENSDART00000131640
|
tiam1a
|
T cell lymphoma invasion and metastasis 1a |
| chr21_-_40782393 | 0.13 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr17_+_23556764 | 0.13 |
ENSDART00000146787
|
pank1a
|
pantothenate kinase 1a |
| chr10_-_36633882 | 0.13 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr19_-_29853402 | 0.13 |
ENSDART00000024292
ENSDART00000188508 |
txlna
|
taxilin alpha |
| chr1_+_29766725 | 0.13 |
ENSDART00000054064
|
zc3h13
|
zinc finger CCCH-type containing 13 |
| chr7_-_69636502 | 0.13 |
ENSDART00000126739
|
tspan5a
|
tetraspanin 5a |
| chr12_+_20122711 | 0.13 |
ENSDART00000184411
|
mgrn1a
|
mahogunin, ring finger 1a |
| chr5_-_30615901 | 0.13 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr17_+_24632440 | 0.13 |
ENSDART00000157092
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr4_+_13953537 | 0.13 |
ENSDART00000133596
|
pphln1
|
periphilin 1 |
| chr3_+_52999962 | 0.12 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr7_-_38340674 | 0.12 |
ENSDART00000075782
|
slc7a10a
|
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10a |
| chr2_+_45068366 | 0.12 |
ENSDART00000142175
|
si:dkey-76d14.2
|
si:dkey-76d14.2 |
| chr12_-_20120702 | 0.12 |
ENSDART00000153387
ENSDART00000158412 ENSDART00000112768 |
ubald1a
|
UBA-like domain containing 1a |
| chr6_-_45949121 | 0.12 |
ENSDART00000058555
|
tardbp
|
TAR DNA binding protein |
| chr8_+_23093155 | 0.12 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr24_-_33756003 | 0.12 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr13_+_28732101 | 0.12 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr8_-_19467011 | 0.12 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr23_-_44226556 | 0.12 |
ENSDART00000149115
|
zgc:158659
|
zgc:158659 |
| chr24_+_7495945 | 0.12 |
ENSDART00000133525
ENSDART00000182460 ENSDART00000162954 |
kmt2ca
|
lysine (K)-specific methyltransferase 2Ca |
| chr23_-_17866100 | 0.11 |
ENSDART00000079506
|
tfip11
|
tuftelin interacting protein 11 |
| chr6_-_40768654 | 0.11 |
ENSDART00000184668
ENSDART00000146470 |
arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr22_+_18886209 | 0.11 |
ENSDART00000144402
|
fstl3
|
follistatin-like 3 (secreted glycoprotein) |
| chr23_+_36083529 | 0.11 |
ENSDART00000053295
ENSDART00000130260 |
hoxc10a
|
homeobox C10a |
| chr22_+_38276024 | 0.11 |
ENSDART00000143792
|
rcor3
|
REST corepressor 3 |
| chr5_-_5669879 | 0.11 |
ENSDART00000191963
|
CABZ01075628.1
|
|
| chr12_+_9817440 | 0.11 |
ENSDART00000137081
ENSDART00000123712 |
rundc3ab
|
RUN domain containing 3Ab |
| chr15_+_28368644 | 0.11 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr4_-_15003854 | 0.11 |
ENSDART00000134701
ENSDART00000002401 |
klhdc10
|
kelch domain containing 10 |
| chr15_+_28368823 | 0.11 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr22_+_4443689 | 0.11 |
ENSDART00000185490
|
ticam1
|
toll-like receptor adaptor molecule 1 |
| chr14_-_12106603 | 0.10 |
ENSDART00000054619
|
prps1b
|
phosphoribosyl pyrophosphate synthetase 1B |
| chr13_-_33317323 | 0.10 |
ENSDART00000110295
ENSDART00000144848 ENSDART00000136701 |
tmem234
|
transmembrane protein 234 |
| chr17_+_19626479 | 0.10 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr17_+_20504196 | 0.10 |
ENSDART00000190539
|
neurl1ab
|
neuralized E3 ubiquitin protein ligase 1Ab |
| chr23_-_3759345 | 0.10 |
ENSDART00000132205
ENSDART00000137707 ENSDART00000189382 |
hmga1a
|
high mobility group AT-hook 1a |
| chr6_+_11249706 | 0.09 |
ENSDART00000186547
ENSDART00000193287 |
atg9a
|
ATG9 autophagy related 9 homolog A (S. cerevisiae) |
| chr8_-_44611357 | 0.09 |
ENSDART00000063396
|
bag4
|
BCL2 associated athanogene 4 |
| chr5_+_23045096 | 0.09 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr15_-_15468326 | 0.09 |
ENSDART00000161192
|
rab34a
|
RAB34, member RAS oncogene family a |
| chr3_+_34985895 | 0.09 |
ENSDART00000121981
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr7_-_31618166 | 0.09 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr7_-_12909352 | 0.09 |
ENSDART00000172901
|
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr11_+_35364445 | 0.09 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr25_-_3503164 | 0.09 |
ENSDART00000191477
ENSDART00000186345 ENSDART00000180199 |
PRKAR2B
|
si:ch211-272n13.7 |
| chr3_-_21348478 | 0.08 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr19_-_19339285 | 0.08 |
ENSDART00000158413
ENSDART00000170479 |
cspg5b
|
chondroitin sulfate proteoglycan 5b |
| chr5_+_41477954 | 0.08 |
ENSDART00000185871
|
pias2
|
protein inhibitor of activated STAT, 2 |
| chr22_+_14117078 | 0.07 |
ENSDART00000013575
|
bzw1a
|
basic leucine zipper and W2 domains 1a |
| chr23_-_31974060 | 0.07 |
ENSDART00000168087
|
BX927210.1
|
|
| chr6_+_18367388 | 0.07 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr13_+_5978809 | 0.07 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr17_-_24680965 | 0.07 |
ENSDART00000154880
|
arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr18_+_30998472 | 0.07 |
ENSDART00000154993
ENSDART00000099333 |
cd151l
|
CD151 antigen, like |
| chr23_+_41800052 | 0.07 |
ENSDART00000141484
|
pdyn
|
prodynorphin |
| chr23_+_9088191 | 0.07 |
ENSDART00000030811
|
cables2b
|
Cdk5 and Abl enzyme substrate 2b |
| chr20_-_26846028 | 0.07 |
ENSDART00000136687
|
mylk4b
|
myosin light chain kinase family, member 4b |
| chr14_-_2933185 | 0.07 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr5_+_19448078 | 0.07 |
ENSDART00000088968
|
ube3b
|
ubiquitin protein ligase E3B |
| chr15_-_22147860 | 0.07 |
ENSDART00000149784
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr14_-_8080416 | 0.07 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox4a+sox4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.1 | 0.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 0.9 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) pyrimidine dimer repair(GO:0006290) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.2 | GO:1903792 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.1 | 0.3 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:0003156 | regulation of organ formation(GO:0003156) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.1 | GO:0042941 | D-amino acid transport(GO:0042940) D-alanine transport(GO:0042941) D-serine transport(GO:0042942) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.4 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:1902165 | regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902165) |
| 0.0 | 0.1 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.0 | 0.2 | GO:0010692 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.7 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.0 | 0.1 | GO:0032816 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) positive regulation of myeloid leukocyte cytokine production involved in immune response(GO:0061081) myeloid leukocyte cytokine production(GO:0061082) |
| 0.0 | 0.2 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.7 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.3 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.3 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.3 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:1903019 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.2 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0030837 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 0.2 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.2 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.3 | GO:0004361 | glutaryl-CoA dehydrogenase activity(GO:0004361) |
| 0.1 | 0.4 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.2 | GO:0032404 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.1 | 0.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 0.2 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.5 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |