Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
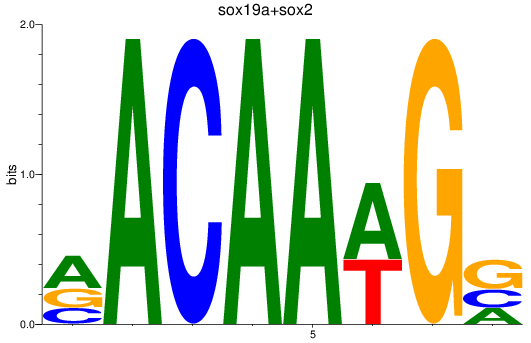
Results for sox19a+sox2
Z-value: 2.32
Transcription factors associated with sox19a+sox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox19a
|
ENSDARG00000010770 | SRY-box transcription factor 19a |
|
sox2
|
ENSDARG00000070913 | SRY-box transcription factor 2 |
|
sox19a
|
ENSDARG00000110497 | SRY-box transcription factor 19a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox2 | dr11_v1_chr22_-_37349967_37349967 | -0.65 | 3.3e-03 | Click! |
| sox19a | dr11_v1_chr5_-_24201437_24201437 | -0.39 | 1.1e-01 | Click! |
Activity profile of sox19a+sox2 motif
Sorted Z-values of sox19a+sox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_2393764 | 8.45 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr5_+_44846434 | 8.17 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr12_+_48340133 | 6.85 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr13_-_17860307 | 5.60 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr14_-_5642371 | 5.11 |
ENSDART00000183859
ENSDART00000054876 |
npm1b
|
nucleophosmin 1b |
| chr20_-_29498178 | 4.91 |
ENSDART00000152986
ENSDART00000027851 ENSDART00000152954 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr1_+_44173506 | 4.91 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr5_-_28149767 | 4.76 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr1_+_44173245 | 4.58 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr12_-_33357655 | 4.52 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr5_+_44846280 | 4.51 |
ENSDART00000084370
|
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr23_+_30736895 | 4.33 |
ENSDART00000042944
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr2_+_38025260 | 4.30 |
ENSDART00000075905
|
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr7_+_58751504 | 4.11 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr11_-_25257595 | 4.05 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr10_+_3153973 | 4.03 |
ENSDART00000183223
|
hic2
|
hypermethylated in cancer 2 |
| chr8_+_52442622 | 4.02 |
ENSDART00000012758
|
zgc:77112
|
zgc:77112 |
| chr10_+_2842923 | 4.00 |
ENSDART00000181895
|
ykt6
|
YKT6 v-SNARE homolog (S. cerevisiae) |
| chr2_-_49031303 | 3.99 |
ENSDART00000143471
|
cdc34b
|
cell division cycle 34 homolog (S. cerevisiae) b |
| chr18_+_8912113 | 3.93 |
ENSDART00000147467
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr4_-_837768 | 3.89 |
ENSDART00000185280
ENSDART00000135618 |
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr9_+_8396755 | 3.89 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr20_+_26939742 | 3.85 |
ENSDART00000138369
ENSDART00000062061 ENSDART00000152992 |
cdca4
|
cell division cycle associated 4 |
| chr5_+_57924611 | 3.78 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr12_-_33354409 | 3.76 |
ENSDART00000178515
|
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr11_-_26832685 | 3.75 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr21_-_30082414 | 3.71 |
ENSDART00000157307
ENSDART00000155188 |
ccnjl
|
cyclin J-like |
| chr13_+_30506781 | 3.67 |
ENSDART00000110884
|
zmiz1a
|
zinc finger, MIZ-type containing 1a |
| chr2_-_32688905 | 3.67 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr16_+_35535171 | 3.63 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr15_+_11644866 | 3.59 |
ENSDART00000188716
|
slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr3_+_19245804 | 3.57 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr15_-_16076399 | 3.56 |
ENSDART00000135658
ENSDART00000133755 ENSDART00000080413 |
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr19_-_47570672 | 3.47 |
ENSDART00000112155
|
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr19_-_47832853 | 3.43 |
ENSDART00000170988
|
ago4
|
argonaute RISC catalytic component 4 |
| chr20_+_26940178 | 3.31 |
ENSDART00000190888
|
cdca4
|
cell division cycle associated 4 |
| chr18_-_25051846 | 3.30 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr11_-_25257045 | 3.25 |
ENSDART00000130477
|
snai1a
|
snail family zinc finger 1a |
| chr8_+_23356264 | 3.22 |
ENSDART00000145062
|
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr5_-_15494164 | 3.16 |
ENSDART00000140668
ENSDART00000188076 ENSDART00000085943 |
taok3a
|
TAO kinase 3a |
| chr18_+_8912710 | 3.12 |
ENSDART00000142866
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr21_-_18275226 | 3.12 |
ENSDART00000126672
ENSDART00000135239 |
brd3a
|
bromodomain containing 3a |
| chr22_-_10541372 | 3.11 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr6_-_49547680 | 3.10 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr1_+_22654875 | 3.10 |
ENSDART00000019698
ENSDART00000161874 |
anxa5b
|
annexin A5b |
| chr19_-_18135724 | 3.08 |
ENSDART00000186609
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr25_+_33063762 | 3.06 |
ENSDART00000189974
|
tln2b
|
talin 2b |
| chr16_+_35535375 | 3.05 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr14_+_6963312 | 3.05 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr18_+_8912536 | 3.03 |
ENSDART00000134827
ENSDART00000061904 |
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr5_-_33230794 | 3.00 |
ENSDART00000144273
|
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr20_-_3238110 | 2.99 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr15_-_31265375 | 2.99 |
ENSDART00000086592
|
vezf1b
|
vascular endothelial zinc finger 1b |
| chr8_+_14987006 | 2.97 |
ENSDART00000045038
|
fnbp1l
|
formin binding protein 1-like |
| chr23_-_36449111 | 2.96 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr6_+_23809501 | 2.93 |
ENSDART00000168701
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr8_+_14986833 | 2.93 |
ENSDART00000171867
ENSDART00000191374 |
fnbp1l
|
formin binding protein 1-like |
| chr3_+_43102010 | 2.92 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr5_-_72390259 | 2.91 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr11_+_13424116 | 2.91 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr19_-_47571456 | 2.90 |
ENSDART00000158071
ENSDART00000165841 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr1_+_49878000 | 2.89 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr20_-_3319642 | 2.88 |
ENSDART00000186743
ENSDART00000123096 |
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr7_-_26601307 | 2.87 |
ENSDART00000188934
|
plscr3b
|
phospholipid scramblase 3b |
| chr17_-_45125537 | 2.87 |
ENSDART00000113552
|
zgc:163014
|
zgc:163014 |
| chr23_-_10745288 | 2.87 |
ENSDART00000140745
ENSDART00000013768 |
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr22_+_15960005 | 2.87 |
ENSDART00000033617
|
stil
|
scl/tal1 interrupting locus |
| chr25_+_8921425 | 2.86 |
ENSDART00000128591
|
accs
|
1-aminocyclopropane-1-carboxylate synthase homolog (Arabidopsis)(non-functional) |
| chr19_-_46037835 | 2.85 |
ENSDART00000163815
|
nup153
|
nucleoporin 153 |
| chr2_+_15048410 | 2.83 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr17_+_1360192 | 2.83 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr22_+_15959844 | 2.82 |
ENSDART00000182201
|
stil
|
scl/tal1 interrupting locus |
| chr4_-_20108833 | 2.82 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr15_-_28587147 | 2.81 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr23_-_31645760 | 2.80 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr21_+_3166988 | 2.78 |
ENSDART00000168163
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr23_-_3758637 | 2.76 |
ENSDART00000131536
ENSDART00000139408 ENSDART00000137826 |
hmga1a
|
high mobility group AT-hook 1a |
| chr7_+_40081630 | 2.75 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr20_+_51730658 | 2.75 |
ENSDART00000010271
|
aida
|
axin interactor, dorsalization associated |
| chr2_-_37477654 | 2.74 |
ENSDART00000193921
|
dapk3
|
death-associated protein kinase 3 |
| chr21_+_34119759 | 2.74 |
ENSDART00000024750
ENSDART00000128242 |
hmgb3b
|
high mobility group box 3b |
| chr12_+_23991639 | 2.74 |
ENSDART00000003143
|
psme4b
|
proteasome activator subunit 4b |
| chr17_-_4252221 | 2.73 |
ENSDART00000152020
|
gdf3
|
growth differentiation factor 3 |
| chr5_-_22602979 | 2.73 |
ENSDART00000146287
|
nono
|
non-POU domain containing, octamer-binding |
| chr3_+_7771420 | 2.72 |
ENSDART00000156809
ENSDART00000156309 |
hook2
|
hook microtubule-tethering protein 2 |
| chr18_+_45573416 | 2.71 |
ENSDART00000132184
ENSDART00000145288 |
kifc3
|
kinesin family member C3 |
| chr12_-_4475890 | 2.71 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr19_-_23249822 | 2.71 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr5_+_33301005 | 2.71 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr22_+_15960514 | 2.70 |
ENSDART00000181617
|
stil
|
scl/tal1 interrupting locus |
| chr18_+_39487486 | 2.70 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr7_+_38380135 | 2.69 |
ENSDART00000174005
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr14_-_7306983 | 2.68 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr22_+_2751887 | 2.67 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr12_-_10508952 | 2.66 |
ENSDART00000152806
|
zgc:152977
|
zgc:152977 |
| chr12_+_23991276 | 2.65 |
ENSDART00000153136
|
psme4b
|
proteasome activator subunit 4b |
| chr2_-_51087077 | 2.64 |
ENSDART00000167987
|
ftr67
|
finTRIM family, member 67 |
| chr8_-_30742233 | 2.64 |
ENSDART00000098986
|
gucd1
|
guanylyl cyclase domain containing 1 |
| chr18_-_7032227 | 2.64 |
ENSDART00000127138
|
calub
|
calumenin b |
| chr6_+_23809163 | 2.63 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr19_+_14352332 | 2.63 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr2_-_21170517 | 2.63 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr10_+_15255012 | 2.62 |
ENSDART00000023766
|
vldlr
|
very low density lipoprotein receptor |
| chr12_-_27212596 | 2.62 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr14_+_12391871 | 2.62 |
ENSDART00000115408
ENSDART00000106626 ENSDART00000054611 ENSDART00000126655 |
thoc2
|
THO complex 2 |
| chr6_-_24066659 | 2.62 |
ENSDART00000164159
ENSDART00000172051 |
dr1
|
down-regulator of transcription 1 |
| chr7_-_53117131 | 2.61 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr9_-_28937880 | 2.60 |
ENSDART00000132878
|
epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr8_+_45338073 | 2.58 |
ENSDART00000185024
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr10_-_36633882 | 2.58 |
ENSDART00000077161
ENSDART00000137688 |
rsf1b.1
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 remodeling and spacing factor 1b, tandem duplicate 1 |
| chr10_-_32663759 | 2.58 |
ENSDART00000126727
|
atg101
|
autophagy related 101 |
| chr10_-_28380919 | 2.58 |
ENSDART00000183409
ENSDART00000183105 ENSDART00000100207 ENSDART00000185392 ENSDART00000131220 |
btg3
|
B-cell translocation gene 3 |
| chr8_+_52442785 | 2.58 |
ENSDART00000189958
|
zgc:77112
|
zgc:77112 |
| chr2_-_34138400 | 2.58 |
ENSDART00000056667
|
cenpl
|
centromere protein L |
| chr1_-_31083535 | 2.57 |
ENSDART00000138113
|
ppp1r9alb
|
protein phosphatase 1 regulatory subunit 9A-like B |
| chr10_+_15255198 | 2.56 |
ENSDART00000139047
ENSDART00000172107 ENSDART00000183413 ENSDART00000185314 |
vldlr
|
very low density lipoprotein receptor |
| chr3_-_32873641 | 2.54 |
ENSDART00000075277
|
zgc:113090
|
zgc:113090 |
| chr8_+_12951155 | 2.53 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr22_-_37565348 | 2.53 |
ENSDART00000149482
ENSDART00000104478 |
fxr1
|
fragile X mental retardation, autosomal homolog 1 |
| chr2_-_26596794 | 2.53 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr10_+_17371356 | 2.52 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr16_-_42175617 | 2.52 |
ENSDART00000084715
|
alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr13_-_35908275 | 2.51 |
ENSDART00000013961
|
mycla
|
MYCL proto-oncogene, bHLH transcription factor a |
| chr2_+_26240339 | 2.51 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr24_-_24724233 | 2.51 |
ENSDART00000127044
ENSDART00000012399 |
armc1
|
armadillo repeat containing 1 |
| chr23_+_9522942 | 2.50 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr9_-_54840124 | 2.48 |
ENSDART00000137214
ENSDART00000085693 |
gpm6bb
|
glycoprotein M6Bb |
| chr11_-_44194132 | 2.48 |
ENSDART00000182954
ENSDART00000111271 |
CABZ01080074.1
|
|
| chr19_+_7636941 | 2.47 |
ENSDART00000081611
ENSDART00000163805 ENSDART00000112404 |
cgnb
|
cingulin b |
| chr2_-_44777592 | 2.47 |
ENSDART00000113351
ENSDART00000169310 |
ncapd2
|
non-SMC condensin I complex, subunit D2 |
| chr5_-_22602780 | 2.47 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr18_-_12957451 | 2.47 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr8_+_30742898 | 2.47 |
ENSDART00000018475
|
snrpd3
|
small nuclear ribonucleoprotein D3 polypeptide |
| chr5_+_40835601 | 2.46 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr18_+_22174630 | 2.45 |
ENSDART00000089549
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr17_+_15535501 | 2.45 |
ENSDART00000002932
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr12_-_17592215 | 2.45 |
ENSDART00000134597
|
usp42
|
ubiquitin specific peptidase 42 |
| chr14_-_24277805 | 2.44 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr8_-_19467011 | 2.43 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr22_-_5171362 | 2.43 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr5_+_26212621 | 2.42 |
ENSDART00000134432
|
oclnb
|
occludin b |
| chr2_+_32846602 | 2.41 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr14_+_15155684 | 2.41 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr9_+_21793565 | 2.40 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr3_-_26191960 | 2.40 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr12_-_27212880 | 2.39 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr21_-_43665537 | 2.39 |
ENSDART00000157610
|
si:dkey-229d11.3
|
si:dkey-229d11.3 |
| chr5_-_3960161 | 2.38 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr20_+_26943072 | 2.37 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr9_+_33340311 | 2.37 |
ENSDART00000140064
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr2_-_10386738 | 2.37 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr6_-_26080384 | 2.36 |
ENSDART00000157181
ENSDART00000154568 |
hs2st1b
|
heparan sulfate 2-O-sulfotransferase 1b |
| chr20_+_27749133 | 2.35 |
ENSDART00000089013
|
vrtn
|
vertebrae development associated |
| chr9_-_41090048 | 2.35 |
ENSDART00000131681
ENSDART00000182552 |
asnsd1
|
asparagine synthetase domain containing 1 |
| chr8_-_1838315 | 2.35 |
ENSDART00000114476
ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr10_-_21362320 | 2.34 |
ENSDART00000189789
|
avd
|
avidin |
| chr21_+_15790366 | 2.34 |
ENSDART00000101956
|
ptpra
|
protein tyrosine phosphatase, receptor type, A |
| chr22_+_336256 | 2.34 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr25_-_12902242 | 2.33 |
ENSDART00000164733
|
sept15
|
septin 15 |
| chr16_+_25259313 | 2.33 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr10_-_35257458 | 2.33 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr22_-_10541712 | 2.33 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr5_+_29831235 | 2.32 |
ENSDART00000109660
|
f11r.1
|
F11 receptor, tandem duplicate 1 |
| chr23_+_36308428 | 2.32 |
ENSDART00000134607
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr23_-_27822920 | 2.32 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr7_-_24875421 | 2.31 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr5_+_3891485 | 2.30 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr5_-_56964547 | 2.28 |
ENSDART00000074400
|
tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr2_+_26240631 | 2.28 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr21_+_12056934 | 2.27 |
ENSDART00000125380
|
zgc:162344
|
zgc:162344 |
| chr8_+_47677208 | 2.27 |
ENSDART00000123254
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr13_-_45022527 | 2.25 |
ENSDART00000159021
|
khdrbs1a
|
KH domain containing, RNA binding, signal transduction associated 1a |
| chr12_+_30788912 | 2.25 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr5_+_34549845 | 2.24 |
ENSDART00000139317
|
aif1l
|
allograft inflammatory factor 1-like |
| chr13_+_51710725 | 2.24 |
ENSDART00000163741
|
pwwp2b
|
PWWP domain containing 2B |
| chr19_+_46113828 | 2.23 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr13_+_22675802 | 2.23 |
ENSDART00000145538
ENSDART00000143312 |
zgc:193505
|
zgc:193505 |
| chr15_+_24756860 | 2.22 |
ENSDART00000156424
ENSDART00000078035 |
cpda
|
carboxypeptidase D, a |
| chr3_+_29082267 | 2.22 |
ENSDART00000145615
|
cacna1i
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr8_-_9570511 | 2.20 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr2_-_21167652 | 2.19 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr7_+_36467796 | 2.19 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr23_+_36308717 | 2.19 |
ENSDART00000131711
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr6_-_46735380 | 2.18 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr16_+_26846495 | 2.18 |
ENSDART00000078124
|
trim35-29
|
tripartite motif containing 35-29 |
| chr5_-_39805620 | 2.18 |
ENSDART00000137801
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr17_+_28005763 | 2.18 |
ENSDART00000155838
|
luzp1
|
leucine zipper protein 1 |
| chr15_+_29025090 | 2.17 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr14_-_32876280 | 2.15 |
ENSDART00000173168
|
si:rp71-46j2.7
|
si:rp71-46j2.7 |
| chr22_-_718615 | 2.15 |
ENSDART00000149320
|
arl8a
|
ADP-ribosylation factor-like 8A |
| chr17_-_24866727 | 2.15 |
ENSDART00000027957
|
hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr16_-_31342356 | 2.14 |
ENSDART00000136648
|
mroh1
|
maestro heat-like repeat family member 1 |
| chr14_+_35464994 | 2.14 |
ENSDART00000115307
|
si:ch211-203d1.3
|
si:ch211-203d1.3 |
| chr11_-_11792766 | 2.13 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr5_+_36666715 | 2.13 |
ENSDART00000097686
|
zgc:153990
|
zgc:153990 |
| chr11_-_24063196 | 2.13 |
ENSDART00000036513
|
trib3
|
tribbles pseudokinase 3 |
| chr3_+_32791786 | 2.12 |
ENSDART00000180174
|
tbc1d10b
|
TBC1 domain family, member 10b |
| chr2_-_23390779 | 2.12 |
ENSDART00000020136
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr2_-_32262287 | 2.12 |
ENSDART00000056621
ENSDART00000039717 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr10_+_11261576 | 2.11 |
ENSDART00000155333
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox19a+sox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.4 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 1.4 | 5.6 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 1.2 | 13.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 1.0 | 15.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.0 | 3.8 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 1.0 | 3.8 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.9 | 8.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.9 | 5.5 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.9 | 5.4 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.9 | 18.5 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.9 | 2.6 | GO:0090134 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.9 | 2.6 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.8 | 2.5 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) negative regulation of anion transport(GO:1903792) |
| 0.8 | 5.6 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.8 | 2.4 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.7 | 2.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.7 | 2.2 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) |
| 0.7 | 2.8 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.7 | 2.0 | GO:0002940 | tRNA N2-guanine methylation(GO:0002940) |
| 0.7 | 2.6 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.6 | 4.5 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.6 | 2.5 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.6 | 2.9 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.6 | 5.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.6 | 1.7 | GO:0019852 | L-ascorbic acid transport(GO:0015882) L-ascorbic acid metabolic process(GO:0019852) |
| 0.6 | 1.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.5 | 1.6 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.5 | 2.7 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.5 | 2.7 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.5 | 2.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.5 | 3.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.5 | 1.5 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.5 | 1.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.5 | 2.0 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.5 | 4.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.5 | 1.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.5 | 1.9 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.5 | 1.4 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.5 | 1.4 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.5 | 2.3 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.4 | 1.3 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.4 | 1.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.4 | 1.3 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.4 | 1.3 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.4 | 1.3 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.4 | 1.3 | GO:0035666 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.4 | 2.1 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.4 | 1.2 | GO:0030237 | female sex determination(GO:0030237) |
| 0.4 | 3.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.4 | 1.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) positive regulation of lamellipodium organization(GO:1902745) |
| 0.4 | 1.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.4 | 4.1 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.4 | 3.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 0.8 | GO:0032196 | transposition(GO:0032196) |
| 0.4 | 0.8 | GO:1990120 | messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.4 | 1.5 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.4 | 1.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 1.1 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.4 | 4.4 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.4 | 1.5 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 0.7 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.4 | 1.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.4 | 8.1 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.4 | 3.5 | GO:1901678 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.3 | 1.0 | GO:0034476 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.3 | 2.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.3 | 3.1 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.3 | 1.7 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.3 | 1.7 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.3 | 5.6 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.3 | 3.0 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.3 | 3.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.3 | 1.3 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.3 | 1.0 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.3 | 1.3 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.3 | 1.2 | GO:1901166 | neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 0.3 | 2.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 1.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.3 | 1.8 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 6.6 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.3 | 1.8 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.3 | 1.5 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.3 | 4.7 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.3 | 1.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.3 | 0.9 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 1.4 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism metabolic process(GO:0044033) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 0.8 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.3 | 2.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.3 | 0.8 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.3 | 1.9 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.3 | 1.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.3 | 1.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.3 | 1.6 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.3 | 2.9 | GO:0039023 | pronephric duct morphogenesis(GO:0039023) |
| 0.3 | 10.5 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.3 | 1.8 | GO:0030728 | ovulation(GO:0030728) |
| 0.3 | 1.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.3 | 1.3 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.3 | 0.8 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.3 | 1.3 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.3 | 1.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.3 | 2.5 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.3 | 1.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) negative regulation of muscle cell differentiation(GO:0051148) |
| 0.3 | 1.3 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.2 | 2.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 1.2 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.2 | 1.2 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.2 | 1.2 | GO:0030328 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.2 | 11.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.2 | 1.9 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.2 | 21.1 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.2 | 1.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.2 | 1.1 | GO:0072575 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.2 | 2.2 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.2 | 0.7 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.2 | 1.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 20.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.2 | 1.1 | GO:0045448 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 2.5 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.2 | 3.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 5.8 | GO:2001243 | negative regulation of intrinsic apoptotic signaling pathway(GO:2001243) |
| 0.2 | 0.2 | GO:0072539 | CD4-positive, alpha-beta T cell differentiation involved in immune response(GO:0002294) T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) T-helper cell differentiation(GO:0042093) CD4-positive, alpha-beta T cell differentiation(GO:0043367) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.2 | 1.0 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.2 | 1.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 0.8 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 1.4 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 1.8 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.2 | 3.5 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 1.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.2 | 2.1 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 1.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.2 | 0.9 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.2 | 0.9 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.2 | 0.9 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.2 | 1.5 | GO:0070897 | DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.2 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 1.6 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.2 | 3.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 1.0 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.2 | 1.0 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.2 | 5.0 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.2 | 5.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.2 | 1.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.0 | GO:1900153 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) positive regulation of mRNA catabolic process(GO:0061014) regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.2 | 0.7 | GO:0032206 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.2 | 1.9 | GO:0006032 | chitin catabolic process(GO:0006032) |
| 0.2 | 3.3 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.2 | 1.8 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 0.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.2 | 0.8 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.2 | 1.8 | GO:0098887 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.2 | 2.1 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.2 | 1.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.2 | 2.4 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 0.2 | 1.1 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 2.2 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 1.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 3.6 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.2 | 1.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.2 | 7.8 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.2 | 1.1 | GO:0090177 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 0.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.2 | 1.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 0.3 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.1 | 1.2 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 5.6 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 1.4 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 12.5 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.1 | 0.7 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.8 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.1 | 1.8 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.7 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 0.5 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.1 | 1.6 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.1 | 0.5 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.8 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 1.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.1 | 3.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 5.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.3 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 1.4 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 0.9 | GO:0098937 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 1.8 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 3.0 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.1 | 0.9 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.1 | 0.7 | GO:1900028 | negative regulation of Rac protein signal transduction(GO:0035021) wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 0.6 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 1.7 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 0.4 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.1 | 1.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.7 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 1.6 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.1 | 0.2 | GO:1904871 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.1 | 2.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 7.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.1 | 0.5 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.6 | GO:0031174 | lifelong otolith mineralization(GO:0031174) |
| 0.1 | 0.4 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.1 | 1.4 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 1.9 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.5 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.1 | 0.8 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 4.4 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.1 | 0.6 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.4 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.1 | 5.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.1 | 2.5 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 2.1 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.1 | 2.4 | GO:0046580 | negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.1 | 1.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.1 | 0.4 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA transport(GO:0051030) snRNA import into nucleus(GO:0061015) |
| 0.1 | 7.4 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.1 | 3.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 13.6 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.1 | 1.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.6 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.1 | 1.1 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.1 | 0.9 | GO:0031268 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.6 | GO:0000730 | DNA recombinase assembly(GO:0000730) |
| 0.1 | 0.5 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.1 | 10.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.1 | 0.9 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) cellular response to dsRNA(GO:0071359) |
| 0.1 | 1.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.5 | GO:0048532 | anatomical structure arrangement(GO:0048532) |
| 0.1 | 0.3 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 1.0 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.1 | 0.6 | GO:0090308 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 1.3 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.1 | 0.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 7.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 1.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.1 | 1.7 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 0.8 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 1.1 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.5 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 1.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.7 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.1 | 1.8 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.4 | GO:0016246 | RNA interference(GO:0016246) |
| 0.1 | 2.0 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.1 | 0.6 | GO:0034063 | cytoplasmic mRNA processing body assembly(GO:0033962) stress granule assembly(GO:0034063) |
| 0.1 | 1.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.1 | 1.3 | GO:0006278 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 1.5 | GO:0043506 | regulation of JUN kinase activity(GO:0043506) |
| 0.1 | 1.1 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.2 | GO:0098534 | centriole assembly(GO:0098534) |
| 0.1 | 0.5 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) regulation of vitamin metabolic process(GO:0030656) regulation of retinoic acid biosynthetic process(GO:1900052) |
| 0.1 | 1.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 3.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.3 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 1.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.8 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 1.9 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.1 | 1.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 1.0 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 0.3 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.5 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.1 | 5.5 | GO:0048846 | axon extension involved in axon guidance(GO:0048846) neuron projection extension involved in neuron projection guidance(GO:1902284) |
| 0.1 | 0.2 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.1 | 1.1 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 1.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 1.4 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.5 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.1 | 0.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 1.4 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.6 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.6 | GO:0048640 | negative regulation of developmental growth(GO:0048640) |
| 0.1 | 4.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 1.7 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0034434 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.1 | 3.8 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.1 | 0.3 | GO:0046632 | alpha-beta T cell differentiation(GO:0046632) |
| 0.1 | 1.9 | GO:0030149 | sphingolipid catabolic process(GO:0030149) membrane lipid catabolic process(GO:0046466) |
| 0.1 | 1.5 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 2.3 | GO:0006073 | glycogen metabolic process(GO:0005977) cellular glucan metabolic process(GO:0006073) glucan metabolic process(GO:0044042) |
| 0.1 | 0.5 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.1 | 1.2 | GO:0032786 | positive regulation of DNA-templated transcription, elongation(GO:0032786) |
| 0.1 | 0.8 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.1 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 0.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.9 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 1.5 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.1 | 3.1 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 2.1 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.1 | 5.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.7 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.1 | 0.9 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 1.1 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.2 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 2.2 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.0 | 0.3 | GO:0048823 | nucleate erythrocyte differentiation(GO:0043363) nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.3 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) regulation of cGMP metabolic process(GO:0030823) positive regulation of cGMP metabolic process(GO:0030825) regulation of cGMP biosynthetic process(GO:0030826) positive regulation of cGMP biosynthetic process(GO:0030828) regulation of guanylate cyclase activity(GO:0031282) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.9 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 1.2 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.0 | 0.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 1.0 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 1.9 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.2 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.9 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.3 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 1.0 | GO:0009214 | cyclic nucleotide catabolic process(GO:0009214) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.7 | GO:0060029 | convergent extension involved in organogenesis(GO:0060029) |
| 0.0 | 0.6 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.2 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 2.0 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 2.6 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.0 | 2.4 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 2.6 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.4 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 1.4 | GO:0007052 | mitotic spindle organization(GO:0007052) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.2 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.0 | 0.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 1.6 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.9 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.4 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 4.8 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.6 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 1.0 | GO:0008154 | actin polymerization or depolymerization(GO:0008154) |
| 0.0 | 0.7 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.2 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.1 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.5 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 3.8 | GO:0051604 | protein maturation(GO:0051604) |
| 0.0 | 1.1 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.2 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 0.2 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 3.9 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.4 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.5 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.8 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 1.5 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.3 | GO:0032481 | positive regulation of type I interferon production(GO:0032481) |
| 0.0 | 0.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.3 | GO:0010906 | regulation of glucose metabolic process(GO:0010906) |
| 0.0 | 0.6 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.2 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 0.3 | GO:0060828 | regulation of canonical Wnt signaling pathway(GO:0060828) |
| 0.0 | 0.4 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) |
| 0.0 | 0.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0072662 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 1.4 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 5.3 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 1.0 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.2 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.0 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.0 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0003096 | renal sodium ion transport(GO:0003096) renal absorption(GO:0070293) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.9 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.6 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 0.1 | GO:2000117 | negative regulation of cysteine-type endopeptidase activity(GO:2000117) |
| 0.0 | 0.1 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.6 | GO:0010942 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.0 | 0.1 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 1.3 | 5.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.9 | 2.6 | GO:0031213 | RSF complex(GO:0031213) |
| 0.8 | 5.9 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.7 | 2.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.7 | 3.4 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.7 | 2.0 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.6 | 2.5 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.6 | 2.5 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.6 | 6.8 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.6 | 4.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.6 | 4.7 | GO:0035060 | brahma complex(GO:0035060) |
| 0.6 | 2.9 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.6 | 1.7 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 2.6 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 1.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.5 | 1.4 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.5 | 1.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.5 | 1.8 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.4 | 1.6 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.4 | 6.6 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.4 | 2.3 | GO:0071012 | U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.4 | 5.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 2.6 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.4 | 2.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.4 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.4 | 1.4 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.4 | 5.0 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) |
| 0.4 | 1.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 1.8 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 1.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.3 | 1.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 2.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.3 | 1.7 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.3 | 1.0 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.3 | 1.6 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.3 | 1.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.3 | 10.9 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 2.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 6.5 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.3 | 0.8 | GO:0089701 | U2AF(GO:0089701) |
| 0.3 | 2.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.3 | 1.5 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 2.0 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.3 | 1.8 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 5.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.3 | 4.8 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 4.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.2 | 2.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 1.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.2 | 1.0 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 7.2 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.2 | 0.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.2 | 1.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 2.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 1.0 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.2 | 3.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.2 | 7.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 1.0 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.2 | 0.6 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.2 | 4.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.6 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 2.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 4.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.1 | 0.8 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 2.0 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 2.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 4.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 13.6 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 0.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.1 | 0.9 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.1 | 1.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.6 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 1.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 9.4 | GO:0001726 | ruffle(GO:0001726) |
| 0.1 | 0.6 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.1 | 1.3 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.1 | 2.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 8.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 3.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.8 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 2.7 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 10.8 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.8 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.1 | 1.8 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 4.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.8 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.1 | 0.4 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.1 | 1.6 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.8 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 1.7 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.8 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 0.9 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 1.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 2.0 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 1.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.7 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 0.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 4.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 1.9 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.1 | 5.7 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 0.2 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 2.2 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 0.5 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 3.8 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.1 | 1.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 2.2 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.1 | 2.4 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 4.5 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.1 | 2.0 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 3.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) anchored component of the cytoplasmic side of the plasma membrane(GO:0098753) |
| 0.0 | 5.4 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.5 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 7.2 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 4.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 9.9 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 1.4 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 4.1 | GO:0000323 | lytic vacuole(GO:0000323) |
| 0.0 | 0.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 6.5 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.3 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.2 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 1.3 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 1.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 13.0 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.5 | GO:0055029 | nuclear DNA-directed RNA polymerase complex(GO:0055029) |
| 0.0 | 1.6 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.7 | 6.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 1.6 | 11.5 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.3 | 5.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 1.3 | 6.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 1.0 | 10.4 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 1.0 | 3.8 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.9 | 2.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.9 | 4.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.8 | 2.5 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.8 | 3.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.8 | 5.6 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.8 | 2.4 | GO:0004394 | heparan sulfate 2-O-sulfotransferase activity(GO:0004394) |
| 0.7 | 2.2 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.7 | 3.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.7 | 8.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.7 | 2.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.6 | 3.1 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.6 | 1.8 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.6 | 4.5 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.5 | 2.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.5 | 3.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.5 | 2.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.5 | 2.0 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.5 | 1.5 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.5 | 2.5 | GO:0004307 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.5 | 1.9 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.5 | 1.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.5 | 2.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.4 | 2.2 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.4 | 1.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.4 | 6.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 3.5 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 1.3 | GO:0003978 | UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.4 | 1.3 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.4 | 3.4 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.4 | 1.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.4 | 3.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 1.2 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.4 | 1.2 | GO:0004113 | 2',3'-cyclic-nucleotide 3'-phosphodiesterase activity(GO:0004113) |
| 0.4 | 5.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.4 | 1.5 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.4 | 1.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.4 | 1.5 | GO:0008117 | sphinganine-1-phosphate aldolase activity(GO:0008117) |
| 0.4 | 1.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.4 | 1.4 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.4 | 1.8 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.4 | 1.4 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.3 | 4.2 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.3 | 1.4 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.3 | 4.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 2.7 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.3 | 2.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.3 | 1.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.3 | 2.0 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.3 | 1.6 | GO:0005009 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.3 | 1.0 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.3 | 1.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.3 | 2.5 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 0.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.3 | 0.9 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.3 | 1.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.3 | 2.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.2 | GO:0047611 | acetylspermidine deacetylase activity(GO:0047611) |
| 0.3 | 1.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.3 | 6.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 6.0 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.3 | 1.7 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.3 | 0.8 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.3 | 2.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.7 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.3 | 0.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 3.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.3 | 1.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.3 | 1.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 2.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.3 | 1.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 2.0 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 2.0 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.2 | 0.7 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.2 | 1.7 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.2 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.2 | 2.6 | GO:0000217 | DNA secondary structure binding(GO:0000217) |
| 0.2 | 1.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 1.9 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.2 | 4.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.2 | 1.9 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.2 | 0.7 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.2 | 2.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.2 | 4.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.2 | 5.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 1.0 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.2 | 1.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 1.4 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 6.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 1.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 2.9 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 1.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.2 | 1.5 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 2.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 13.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.2 | 5.5 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 1.6 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.2 | 1.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 1.9 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.8 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.2 | 1.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.2 | 1.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.2 | 0.5 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.2 | 0.6 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.2 | 0.8 | GO:0004122 | cystathionine beta-synthase activity(GO:0004122) |
| 0.2 | 1.2 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.2 | 5.9 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.2 | 2.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 28.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.1 | 0.6 | GO:0051717 | inositol-1,3,4,5-tetrakisphosphate 3-phosphatase activity(GO:0051717) phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.1 | 2.0 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.1 | 2.7 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 3.8 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 3.0 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.7 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.1 | 3.0 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 8.2 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.1 | 0.5 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 1.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.1 | 0.5 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 0.6 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.1 | 0.7 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.6 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 1.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 3.6 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 0.3 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.7 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 1.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 2.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.8 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.1 | 1.3 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.5 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.1 | 1.4 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.1 | 1.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.4 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 1.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.4 | GO:0046404 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.1 | 1.8 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.1 | 1.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.6 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 2.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 4.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 2.1 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.8 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.1 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.9 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.7 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.1 | 0.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 4.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 11.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 7.6 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.2 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.8 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.6 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.1 | 0.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 7.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 1.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 1.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 3.8 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.1 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.4 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 3.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 2.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.1 | 4.6 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.1 | 1.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 2.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.3 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 1.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.3 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 1.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.4 | GO:0034057 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.1 | 0.6 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 21.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.9 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.1 | GO:0008170 | N-methyltransferase activity(GO:0008170) |
| 0.1 | 0.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 2.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.1 | 1.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.8 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.1 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 12.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 0.6 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.6 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 1.5 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 15.9 | GO:0000989 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.4 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 1.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.1 | 2.3 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.6 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.0 | 0.2 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.8 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) protein complex scaffold(GO:0032947) |
| 0.0 | 1.9 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 2.4 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.4 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 1.6 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.7 | GO:0016896 | exoribonuclease activity(GO:0004532) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 2.3 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 1.1 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.6 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.7 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 2.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.0 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.9 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.5 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.2 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.0 | 0.1 | GO:0016521 | pituitary adenylate cyclase activating polypeptide activity(GO:0016521) |
| 0.0 | 1.8 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.4 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 1.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 2.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 2.9 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 2.8 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 2.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.9 | GO:0015211 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine ribonucleotide transmembrane transporter activity(GO:0005346) ATP transmembrane transporter activity(GO:0005347) purine nucleoside transmembrane transporter activity(GO:0015211) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.8 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.2 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 2.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 2.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.4 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0030975 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 13.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type I receptor activity(GO:0001596) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.4 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 2.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.4 | GO:0004518 | nuclease activity(GO:0004518) |
| 0.0 | 3.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 3.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.1 | GO:0033781 | cholesterol 24-hydroxylase activity(GO:0033781) |
| 0.0 | 0.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 2.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 13.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.5 | 7.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 7.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.4 | 5.1 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.3 | 5.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.2 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.2 | 3.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.2 | 12.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.2 | 3.7 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 8.2 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 1.7 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 2.0 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.2 | 2.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 1.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 1.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.1 | 3.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 3.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.6 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 1.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 2.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 3.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.1 | 1.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.1 | 1.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 7.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.2 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 2.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 1.6 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 1.7 | PID MYC PATHWAY | C-MYC pathway |
| 0.1 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 0.9 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.1 | 2.3 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.1 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 3.9 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 1.4 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 1.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.9 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.6 | 3.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.5 | 8.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 13.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.5 | 10.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.4 | 4.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 2.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.3 | 3.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.3 | 2.9 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.3 | 2.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.3 | 1.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 2.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.3 | 2.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 5.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 3.0 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.2 | 3.4 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.2 | 0.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.2 | 0.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.2 | 2.1 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 2.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 0.7 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.2 | 3.8 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.2 | 2.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.2 | 18.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.2 | 2.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.2 | 3.7 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 1.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.2 | 1.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 3.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.1 | 1.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.1 | 1.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.5 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 7.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.7 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.3 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.1 | 4.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 2.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 1.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.0 | REACTOME BASE EXCISION REPAIR | Genes involved in Base Excision Repair |
| 0.1 | 1.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.1 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 2.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 0.6 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 1.5 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.1 | 2.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.1 | 0.9 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.9 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.1 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 2.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 1.2 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 3.3 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.1 | 1.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 2.1 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.1 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 0.8 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 1.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 1.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 1.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 1.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.5 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.1 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.9 | REACTOME PI METABOLISM | Genes involved in PI Metabolism |
| 0.0 | 4.2 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 2.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 4.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.3 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.1 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.3 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 1.0 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |