Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
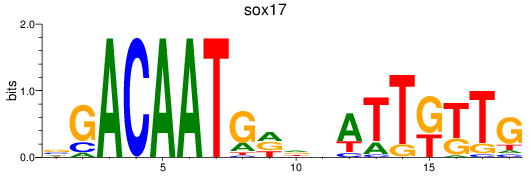
Results for sox17
Z-value: 0.52
Transcription factors associated with sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox17
|
ENSDARG00000101717 | SRY-box transcription factor 17 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox17 | dr11_v1_chr7_-_58776400_58776400 | 0.08 | 7.5e-01 | Click! |
Activity profile of sox17 motif
Sorted Z-values of sox17 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_31854830 | 0.65 |
ENSDART00000148550
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr12_-_3077395 | 0.54 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_41717063 | 0.54 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr5_-_67349916 | 0.53 |
ENSDART00000144092
|
mlxip
|
MLX interacting protein |
| chr7_+_38349667 | 0.52 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr7_-_39378903 | 0.51 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr3_-_40976288 | 0.51 |
ENSDART00000193553
|
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr11_-_44979281 | 0.50 |
ENSDART00000190972
|
ldb1b
|
LIM-domain binding 1b |
| chr25_+_3104959 | 0.50 |
ENSDART00000167130
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr15_-_47468085 | 0.49 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr9_+_38457806 | 0.47 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr7_+_66884570 | 0.47 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr15_-_37543591 | 0.47 |
ENSDART00000180400
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr20_+_4222357 | 0.46 |
ENSDART00000188331
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr15_-_37543839 | 0.46 |
ENSDART00000148870
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr11_-_27537593 | 0.45 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr2_+_51818039 | 0.45 |
ENSDART00000170353
|
acvr2bb
|
activin A receptor type 2Bb |
| chr20_+_4221978 | 0.45 |
ENSDART00000171898
|
ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr19_-_18135724 | 0.44 |
ENSDART00000186609
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr9_-_16218097 | 0.44 |
ENSDART00000190503
|
myo1b
|
myosin IB |
| chr20_-_18794789 | 0.44 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr3_-_40976463 | 0.42 |
ENSDART00000128450
ENSDART00000018676 |
cyp3c1
|
cytochrome P450, family 3, subfamily c, polypeptide 1 |
| chr22_+_12361317 | 0.42 |
ENSDART00000189963
ENSDART00000159614 |
r3hdm1
|
R3H domain containing 1 |
| chr22_+_12361489 | 0.40 |
ENSDART00000182483
|
r3hdm1
|
R3H domain containing 1 |
| chr13_-_35765028 | 0.39 |
ENSDART00000157391
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr4_-_3145359 | 0.39 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_+_30922947 | 0.38 |
ENSDART00000184060
|
cldni
|
claudin i |
| chr3_+_35005730 | 0.37 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr6_+_30533504 | 0.37 |
ENSDART00000155842
|
wwc3
|
WWC family member 3 |
| chr17_-_23895026 | 0.37 |
ENSDART00000122108
|
pdzd8
|
PDZ domain containing 8 |
| chr16_+_50741154 | 0.37 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr21_-_2158298 | 0.37 |
ENSDART00000182199
|
gb:ai877918
|
expressed sequence AI877918 |
| chr10_-_3258073 | 0.36 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr23_+_1414893 | 0.36 |
ENSDART00000173020
|
utrn
|
utrophin |
| chr1_+_53321878 | 0.36 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr10_-_8434816 | 0.34 |
ENSDART00000108643
|
tctn1
|
tectonic family member 1 |
| chr18_+_38807239 | 0.33 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr6_+_4299164 | 0.33 |
ENSDART00000159759
|
nbeal1
|
neurobeachin-like 1 |
| chr2_+_36007449 | 0.33 |
ENSDART00000161837
|
lamc2
|
laminin, gamma 2 |
| chr12_-_7253270 | 0.32 |
ENSDART00000035762
|
ube2d1b
|
ubiquitin-conjugating enzyme E2D 1b |
| chr5_-_69004007 | 0.32 |
ENSDART00000137443
|
ank1a
|
ankyrin 1, erythrocytic a |
| chr5_+_53691803 | 0.32 |
ENSDART00000163256
|
taf6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr1_-_46650022 | 0.32 |
ENSDART00000148759
ENSDART00000053222 |
rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr19_-_25114701 | 0.32 |
ENSDART00000149035
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_+_23147218 | 0.32 |
ENSDART00000030920
ENSDART00000141175 ENSDART00000146264 |
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr23_+_30730121 | 0.31 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr23_+_24973773 | 0.31 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr19_+_46222428 | 0.31 |
ENSDART00000183984
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr20_+_40457599 | 0.31 |
ENSDART00000017553
|
serinc1
|
serine incorporator 1 |
| chr5_+_51833305 | 0.31 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr10_+_15454745 | 0.30 |
ENSDART00000129441
ENSDART00000123935 ENSDART00000163446 ENSDART00000087680 ENSDART00000193752 |
erbin
|
erbb2 interacting protein |
| chr5_+_51833132 | 0.30 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr10_-_7988396 | 0.30 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr16_-_28658341 | 0.30 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr15_+_20344670 | 0.30 |
ENSDART00000158986
|
plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr6_-_7107868 | 0.30 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr2_+_26240631 | 0.29 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr23_-_24542156 | 0.29 |
ENSDART00000132265
|
atp13a2
|
ATPase 13A2 |
| chr8_-_44611357 | 0.29 |
ENSDART00000063396
|
bag4
|
BCL2 associated athanogene 4 |
| chr21_+_6328801 | 0.29 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr5_-_68495967 | 0.29 |
ENSDART00000188107
|
ephb4a
|
eph receptor B4a |
| chr21_+_12036238 | 0.29 |
ENSDART00000102463
ENSDART00000155426 |
zgc:162344
|
zgc:162344 |
| chr4_-_2975461 | 0.29 |
ENSDART00000150794
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr6_+_55428924 | 0.29 |
ENSDART00000018270
|
ncoa5
|
nuclear receptor coactivator 5 |
| chr19_+_27970292 | 0.29 |
ENSDART00000103887
ENSDART00000145549 |
med10
|
mediator complex subunit 10 |
| chr18_-_46183462 | 0.28 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr2_+_22409249 | 0.28 |
ENSDART00000182915
|
zgc:56628
|
zgc:56628 |
| chr13_-_31687925 | 0.28 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr22_-_14161309 | 0.28 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr24_-_16979728 | 0.28 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr5_-_15264007 | 0.27 |
ENSDART00000180641
|
gnb1l
|
guanine nucleotide binding protein (G protein), beta polypeptide 1-like |
| chr5_+_36896933 | 0.27 |
ENSDART00000151984
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr17_+_13031497 | 0.27 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr15_-_30984804 | 0.27 |
ENSDART00000157005
|
nf1a
|
neurofibromin 1a |
| chr20_+_18945057 | 0.27 |
ENSDART00000035447
|
mtmr9
|
myotubularin related protein 9 |
| chr8_+_23147609 | 0.27 |
ENSDART00000180284
|
gid8a
|
GID complex subunit 8 homolog a (S. cerevisiae) |
| chr13_+_31687973 | 0.27 |
ENSDART00000076479
|
slc38a6
|
solute carrier family 38, member 6 |
| chr12_+_29240124 | 0.27 |
ENSDART00000053761
ENSDART00000130172 |
bms1
|
BMS1 ribosome biogenesis factor |
| chr7_-_30624435 | 0.26 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr13_+_18659325 | 0.26 |
ENSDART00000147905
ENSDART00000188249 ENSDART00000134740 |
selenou1a
|
selenoprotein U 1a |
| chr12_-_611367 | 0.26 |
ENSDART00000152286
|
wu:fj29h11
|
wu:fj29h11 |
| chr15_-_28904371 | 0.26 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr17_+_50127648 | 0.26 |
ENSDART00000156460
|
galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr13_-_9367647 | 0.26 |
ENSDART00000083362
ENSDART00000144146 |
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr14_+_16036139 | 0.26 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr3_-_39363065 | 0.26 |
ENSDART00000155894
|
arhgap23a
|
Rho GTPase activating protein 23a |
| chr12_-_30032188 | 0.26 |
ENSDART00000042514
|
atrnl1b
|
attractin-like 1b |
| chr10_-_28513861 | 0.25 |
ENSDART00000177781
|
bbx
|
bobby sox homolog (Drosophila) |
| chr15_-_2803313 | 0.25 |
ENSDART00000060839
|
tgfb1a
|
transforming growth factor, beta 1a |
| chr1_+_46405294 | 0.25 |
ENSDART00000143228
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr23_+_16774589 | 0.25 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr4_-_3064101 | 0.25 |
ENSDART00000135701
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr5_+_24086227 | 0.25 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr19_-_20446756 | 0.24 |
ENSDART00000140711
|
tbc1d5
|
TBC1 domain family, member 5 |
| chr13_-_533243 | 0.24 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr17_+_24222190 | 0.24 |
ENSDART00000181698
ENSDART00000189411 |
ehbp1
|
EH domain binding protein 1 |
| chr7_+_29962559 | 0.24 |
ENSDART00000075538
|
fbxo22
|
F-box protein 22 |
| chr7_-_24204200 | 0.24 |
ENSDART00000087298
|
gmpr2
|
guanosine monophosphate reductase 2 |
| chr10_-_6588793 | 0.24 |
ENSDART00000163788
|
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr4_-_18416566 | 0.24 |
ENSDART00000033717
|
cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr1_+_46404968 | 0.24 |
ENSDART00000042064
|
tubgcp3
|
tubulin, gamma complex associated protein 3 |
| chr25_+_23358924 | 0.24 |
ENSDART00000156965
|
osbpl5
|
oxysterol binding protein-like 5 |
| chr21_+_24536233 | 0.23 |
ENSDART00000145448
ENSDART00000109886 ENSDART00000135191 |
anapc13
|
anaphase promoting complex subunit 13 |
| chr20_-_31238313 | 0.23 |
ENSDART00000028471
|
hpcal1
|
hippocalcin-like 1 |
| chr18_-_21725805 | 0.23 |
ENSDART00000182185
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr18_+_20560616 | 0.23 |
ENSDART00000136710
ENSDART00000151974 ENSDART00000121699 ENSDART00000040074 |
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr11_-_22361306 | 0.23 |
ENSDART00000180688
ENSDART00000182200 ENSDART00000006580 |
tfeb
|
transcription factor EB |
| chr11_+_30636351 | 0.22 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr7_-_28641522 | 0.22 |
ENSDART00000182071
|
slc7a6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
| chr19_-_24136233 | 0.22 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr10_-_22845485 | 0.22 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr18_-_21725638 | 0.21 |
ENSDART00000089853
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr8_-_22326073 | 0.21 |
ENSDART00000084965
|
cep104
|
centrosomal protein 104 |
| chr13_-_48431766 | 0.21 |
ENSDART00000159688
ENSDART00000171765 |
fbxo11a
|
F-box protein 11a |
| chr24_+_41281265 | 0.21 |
ENSDART00000156920
ENSDART00000155151 |
nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr4_-_28353538 | 0.21 |
ENSDART00000064219
|
trmu
|
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr7_-_40122139 | 0.21 |
ENSDART00000173982
|
si:ch73-174h16.5
|
si:ch73-174h16.5 |
| chr21_+_33503835 | 0.20 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr23_-_36670369 | 0.20 |
ENSDART00000006881
|
zbtb39
|
zinc finger and BTB domain containing 39 |
| chr1_+_49878000 | 0.20 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr22_+_1868983 | 0.20 |
ENSDART00000160135
|
si:dkey-15h8.17
|
si:dkey-15h8.17 |
| chr22_+_28818291 | 0.20 |
ENSDART00000136032
|
tp53bp2b
|
tumor protein p53 binding protein, 2b |
| chr17_-_13070602 | 0.20 |
ENSDART00000188311
ENSDART00000193428 |
CU469462.1
|
|
| chr7_+_10562118 | 0.20 |
ENSDART00000185188
ENSDART00000168801 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr6_-_8392104 | 0.20 |
ENSDART00000081561
ENSDART00000181178 |
ilf3a
|
interleukin enhancer binding factor 3a |
| chr17_+_16423721 | 0.20 |
ENSDART00000064233
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr2_+_43204919 | 0.19 |
ENSDART00000160077
ENSDART00000018729 ENSDART00000129134 ENSDART00000056402 |
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr21_-_7178348 | 0.19 |
ENSDART00000187467
|
fam69b
|
family with sequence similarity 69, member B |
| chr18_+_1837668 | 0.19 |
ENSDART00000164210
|
CABZ01079192.1
|
|
| chr7_-_64770456 | 0.19 |
ENSDART00000192618
|
zdhhc21
|
zinc finger, DHHC-type containing 21 |
| chr12_+_19958845 | 0.19 |
ENSDART00000193248
|
ercc4
|
excision repair cross-complementation group 4 |
| chr8_-_9684872 | 0.19 |
ENSDART00000132158
|
ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr17_-_5769196 | 0.18 |
ENSDART00000113885
|
si:dkey-100n19.2
|
si:dkey-100n19.2 |
| chr19_+_42886413 | 0.18 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr3_-_39190317 | 0.18 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr11_+_42494531 | 0.18 |
ENSDART00000067604
|
arf4a
|
ADP-ribosylation factor 4a |
| chr1_+_29766725 | 0.18 |
ENSDART00000054064
|
zc3h13
|
zinc finger CCCH-type containing 13 |
| chr16_-_27138478 | 0.17 |
ENSDART00000147438
|
tmem245
|
transmembrane protein 245 |
| chr10_-_28027490 | 0.17 |
ENSDART00000185445
|
ints2
|
integrator complex subunit 2 |
| chr1_-_23110740 | 0.17 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr6_-_1591002 | 0.17 |
ENSDART00000087039
|
zgc:123305
|
zgc:123305 |
| chr23_+_32101361 | 0.17 |
ENSDART00000138849
|
zgc:56699
|
zgc:56699 |
| chr23_+_25201077 | 0.17 |
ENSDART00000136675
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr21_-_20381481 | 0.16 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr16_+_35905031 | 0.16 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr25_-_31907590 | 0.16 |
ENSDART00000149707
ENSDART00000149471 |
eif3ja
|
eukaryotic translation initiation factor 3, subunit Ja |
| chr10_+_36662640 | 0.16 |
ENSDART00000063359
|
ucp2
|
uncoupling protein 2 |
| chr5_-_8602937 | 0.16 |
ENSDART00000157765
|
rictora
|
RPTOR independent companion of MTOR, complex 2 a |
| chr6_+_54498220 | 0.15 |
ENSDART00000103282
|
si:ch211-233f11.5
|
si:ch211-233f11.5 |
| chr14_+_29609245 | 0.15 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr9_+_16854121 | 0.15 |
ENSDART00000110866
|
cln5
|
CLN5, intracellular trafficking protein |
| chr11_-_42134968 | 0.15 |
ENSDART00000187115
|
FP325130.1
|
|
| chr2_+_55199721 | 0.15 |
ENSDART00000016143
|
zmp:0000000521
|
zmp:0000000521 |
| chr17_+_51520073 | 0.15 |
ENSDART00000189646
ENSDART00000170951 ENSDART00000189492 |
pxdn
|
peroxidasin |
| chr9_-_46276626 | 0.15 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr24_-_33284945 | 0.14 |
ENSDART00000155429
ENSDART00000112845 |
zgc:195173
|
zgc:195173 |
| chr14_+_6954579 | 0.14 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr11_-_30636163 | 0.14 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr25_+_20694177 | 0.14 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr5_-_20678300 | 0.14 |
ENSDART00000088639
|
wscd2
|
WSC domain containing 2 |
| chr16_+_28764017 | 0.14 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr1_-_44704261 | 0.14 |
ENSDART00000133210
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr25_-_7686201 | 0.14 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr2_+_24867534 | 0.14 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr13_+_6189203 | 0.13 |
ENSDART00000109665
|
ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr9_+_21146862 | 0.13 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr16_+_25074029 | 0.13 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr6_+_6780873 | 0.13 |
ENSDART00000011865
|
sec23b
|
Sec23 homolog B, COPII coat complex component |
| chr2_-_26596794 | 0.13 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr6_-_12314475 | 0.13 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr14_-_25956804 | 0.13 |
ENSDART00000135627
ENSDART00000146022 ENSDART00000039660 |
sparc
|
secreted protein, acidic, cysteine-rich (osteonectin) |
| chr4_+_25693463 | 0.12 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr7_+_23495986 | 0.12 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr5_-_33769211 | 0.12 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr21_-_39628771 | 0.12 |
ENSDART00000183995
|
aldocb
|
aldolase C, fructose-bisphosphate, b |
| chr5_-_69314495 | 0.12 |
ENSDART00000182335
|
smtnb
|
smoothelin b |
| chr16_+_28994709 | 0.12 |
ENSDART00000088023
|
gon4l
|
gon-4-like (C. elegans) |
| chr5_+_59278193 | 0.12 |
ENSDART00000160025
|
rasa4
|
RAS p21 protein activator 4 |
| chr18_+_31117323 | 0.12 |
ENSDART00000059893
|
tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr5_-_54790923 | 0.12 |
ENSDART00000176835
|
pik3r1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr22_-_11493236 | 0.12 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr18_+_619619 | 0.12 |
ENSDART00000159846
|
prtga
|
protogenin homolog a (Gallus gallus) |
| chr24_+_26997798 | 0.11 |
ENSDART00000089506
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr13_-_45201300 | 0.11 |
ENSDART00000074750
ENSDART00000180265 |
runx3
|
runt-related transcription factor 3 |
| chr20_+_28803977 | 0.11 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr18_+_31117136 | 0.11 |
ENSDART00000138403
|
tcf25
|
transcription factor 25 (basic helix-loop-helix) |
| chr7_-_32020100 | 0.11 |
ENSDART00000185433
|
kif18a
|
kinesin family member 18A |
| chr1_-_656693 | 0.11 |
ENSDART00000170483
ENSDART00000166786 |
appa
|
amyloid beta (A4) precursor protein a |
| chr3_+_18795570 | 0.10 |
ENSDART00000042368
|
fahd1
|
fumarylacetoacetate hydrolase domain containing 1 |
| chr20_+_32224405 | 0.10 |
ENSDART00000062993
ENSDART00000147448 |
sesn1
|
sestrin 1 |
| chr9_-_32813177 | 0.10 |
ENSDART00000012694
|
mxc
|
myxovirus (influenza virus) resistance C |
| chr1_-_18772241 | 0.10 |
ENSDART00000147645
ENSDART00000133194 ENSDART00000147525 ENSDART00000191493 |
tbc1d1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr25_+_19485198 | 0.10 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr25_-_21894706 | 0.10 |
ENSDART00000189158
|
fbxo31
|
F-box protein 31 |
| chr8_+_10339869 | 0.10 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr14_+_36246726 | 0.10 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr20_-_48898371 | 0.10 |
ENSDART00000170617
|
xrn2
|
5'-3' exoribonuclease 2 |
| chr4_+_9030609 | 0.10 |
ENSDART00000154399
|
aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr6_-_43092175 | 0.10 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr7_-_35314347 | 0.10 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox17
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.3 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.1 | 0.2 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.3 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.5 | GO:0090024 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.1 | 0.2 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.1 | 0.2 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.1 | 0.2 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.5 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.2 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 0.3 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.3 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.3 | GO:0035475 | angioblast cell migration involved in selective angioblast sprouting(GO:0035475) |
| 0.0 | 0.2 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.9 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.3 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate metabolic process(GO:0009256) 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.2 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.6 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.3 | GO:0048696 | collateral sprouting in absence of injury(GO:0048669) regulation of collateral sprouting in absence of injury(GO:0048696) |
| 0.0 | 0.1 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.5 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0000479 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.3 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.0 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.1 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.5 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.1 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:2000651 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.1 | 0.5 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.5 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.4 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.1 | 0.3 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.3 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:1990071 | TRAPPII protein complex(GO:1990071) |
| 0.0 | 0.5 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 1.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.8 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 0.5 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.9 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.3 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.3 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.2 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.9 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.3 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.3 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.9 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |