Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
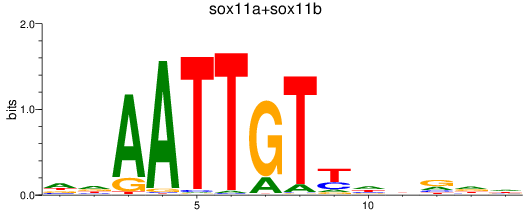
Results for sox11a+sox11b
Z-value: 0.80
Transcription factors associated with sox11a+sox11b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox11a
|
ENSDARG00000077811 | SRY-box transcription factor 11a |
|
sox11b
|
ENSDARG00000095743 | SRY-box transcription factor 11b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox11b | dr11_v1_chr20_-_30035326_30035326 | -0.79 | 1.1e-04 | Click! |
| sox11a | dr11_v1_chr17_-_35881841_35881841 | 0.71 | 9.4e-04 | Click! |
Activity profile of sox11a+sox11b motif
Sorted Z-values of sox11a+sox11b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_3118346 | 1.51 |
ENSDART00000167554
|
hsf5
|
heat shock transcription factor family member 5 |
| chrM_+_6425 | 1.44 |
ENSDART00000093606
|
mt-co1
|
cytochrome c oxidase I, mitochondrial |
| chr10_+_2587234 | 1.25 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr8_+_34434345 | 1.09 |
ENSDART00000190246
ENSDART00000189447 ENSDART00000185557 ENSDART00000189230 |
zgc:174461
|
zgc:174461 |
| chr5_+_66353750 | 1.06 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr23_+_12198242 | 1.03 |
ENSDART00000144104
|
sycp2
|
synaptonemal complex protein 2 |
| chr12_-_19279103 | 0.92 |
ENSDART00000186669
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr20_+_33519435 | 0.92 |
ENSDART00000061829
|
drc1
|
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
| chrM_+_12897 | 0.91 |
ENSDART00000093622
|
mt-nd5
|
NADH dehydrogenase 5, mitochondrial |
| chr16_-_23346095 | 0.89 |
ENSDART00000160546
|
si:dkey-247k7.2
|
si:dkey-247k7.2 |
| chr7_-_22981796 | 0.89 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr13_+_24755049 | 0.88 |
ENSDART00000134102
|
col17a1b
|
collagen, type XVII, alpha 1b |
| chr6_-_57539141 | 0.84 |
ENSDART00000156967
|
itcha
|
itchy E3 ubiquitin protein ligase a |
| chr11_-_25734417 | 0.83 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr2_-_1569250 | 0.83 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr16_+_54263921 | 0.81 |
ENSDART00000002856
|
drd2l
|
dopamine receptor D2 like |
| chr7_-_40145097 | 0.79 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr7_-_58729894 | 0.78 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr18_+_5547185 | 0.72 |
ENSDART00000193977
|
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr13_+_13693722 | 0.72 |
ENSDART00000110509
|
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr2_+_16780643 | 0.71 |
ENSDART00000125647
ENSDART00000108611 ENSDART00000181245 ENSDART00000163194 |
tfa
|
transferrin-a |
| chr11_-_25733910 | 0.69 |
ENSDART00000171935
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr10_+_6884123 | 0.67 |
ENSDART00000149095
ENSDART00000148772 ENSDART00000149334 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr21_-_41305748 | 0.67 |
ENSDART00000170457
|
nsg2
|
neuronal vesicle trafficking associated 2 |
| chr19_+_26718074 | 0.64 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr5_+_41996889 | 0.64 |
ENSDART00000097580
|
pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr10_+_6884627 | 0.63 |
ENSDART00000125262
ENSDART00000121729 ENSDART00000105384 |
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr25_-_3647277 | 0.62 |
ENSDART00000166363
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr25_-_7764083 | 0.62 |
ENSDART00000179800
ENSDART00000181858 |
phf21ab
|
PHD finger protein 21Ab |
| chr10_+_41668483 | 0.60 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr20_+_26538137 | 0.60 |
ENSDART00000045397
|
stx11b.1
|
syntaxin 11b, tandem duplicate 1 |
| chr14_+_4151379 | 0.60 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr11_-_33612433 | 0.59 |
ENSDART00000179756
|
lims1
|
LIM and senescent cell antigen-like domains 1 |
| chr2_+_9061885 | 0.59 |
ENSDART00000028906
|
pigk
|
phosphatidylinositol glycan anchor biosynthesis, class K |
| chr1_-_20928772 | 0.57 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr3_+_26267725 | 0.53 |
ENSDART00000131288
|
adap2
|
ArfGAP with dual PH domains 2 |
| chr18_-_43884044 | 0.52 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr5_+_69716458 | 0.52 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr23_+_31815423 | 0.50 |
ENSDART00000075730
ENSDART00000075726 |
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr3_-_23643751 | 0.49 |
ENSDART00000078425
ENSDART00000140264 |
eve1
|
even-skipped-like1 |
| chr2_-_40135942 | 0.48 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr10_-_28761454 | 0.46 |
ENSDART00000129400
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr15_-_45510977 | 0.46 |
ENSDART00000090596
|
fgf12b
|
fibroblast growth factor 12b |
| chr16_+_18535618 | 0.43 |
ENSDART00000021596
|
rxrbb
|
retinoid x receptor, beta b |
| chr4_-_18635005 | 0.43 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr1_+_26445615 | 0.41 |
ENSDART00000180810
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr24_-_41220538 | 0.41 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr9_+_22657221 | 0.41 |
ENSDART00000101765
|
si:dkey-189g17.2
|
si:dkey-189g17.2 |
| chr25_+_19041329 | 0.40 |
ENSDART00000153467
|
lrtm2b
|
leucine-rich repeats and transmembrane domains 2b |
| chr16_-_43233509 | 0.39 |
ENSDART00000025877
|
cldn12
|
claudin 12 |
| chr24_+_26345609 | 0.39 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
| chr10_-_41906609 | 0.38 |
ENSDART00000102530
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr2_-_23411368 | 0.38 |
ENSDART00000159495
|
si:ch73-129a22.11
|
si:ch73-129a22.11 |
| chr1_+_26444986 | 0.37 |
ENSDART00000046376
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr19_-_31402429 | 0.37 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr12_-_37734973 | 0.36 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| chr1_+_21309386 | 0.36 |
ENSDART00000054442
ENSDART00000145661 |
med28
|
mediator complex subunit 28 |
| chr7_+_22981909 | 0.35 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr12_-_11258404 | 0.35 |
ENSDART00000149229
|
si:ch73-30l9.1
|
si:ch73-30l9.1 |
| chr10_-_41907213 | 0.34 |
ENSDART00000167004
|
kdm2bb
|
lysine (K)-specific demethylase 2Bb |
| chr16_+_36768674 | 0.34 |
ENSDART00000169208
ENSDART00000180470 |
si:ch73-215d9.1
|
si:ch73-215d9.1 |
| chr23_+_23022790 | 0.34 |
ENSDART00000189843
|
samd11
|
sterile alpha motif domain containing 11 |
| chr13_-_45201300 | 0.33 |
ENSDART00000074750
ENSDART00000180265 |
runx3
|
runt-related transcription factor 3 |
| chr15_-_2841677 | 0.33 |
ENSDART00000026145
ENSDART00000180290 |
AMOTL1
|
angiomotin like 1 |
| chr19_-_18135724 | 0.33 |
ENSDART00000186609
|
cbx3a
|
chromobox homolog 3a (HP1 gamma homolog, Drosophila) |
| chr24_-_38657683 | 0.32 |
ENSDART00000154843
|
si:ch1073-164k15.3
|
si:ch1073-164k15.3 |
| chr20_-_31067306 | 0.32 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr16_-_21140097 | 0.32 |
ENSDART00000145837
ENSDART00000146500 |
si:dkey-271j15.3
|
si:dkey-271j15.3 |
| chr22_+_37902976 | 0.32 |
ENSDART00000169327
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr19_-_10915898 | 0.31 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr5_-_26118855 | 0.31 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr5_-_38342992 | 0.30 |
ENSDART00000140337
|
mink1
|
misshapen-like kinase 1 |
| chr22_+_10440991 | 0.30 |
ENSDART00000064805
|
cenpp
|
centromere protein P |
| chr5_-_24869213 | 0.30 |
ENSDART00000112287
ENSDART00000144635 |
gas2l1
|
growth arrest-specific 2 like 1 |
| chr5_-_67911111 | 0.30 |
ENSDART00000051833
|
gsx1
|
GS homeobox 1 |
| chr23_-_31403668 | 0.29 |
ENSDART00000147498
|
CR759830.1
|
|
| chr16_-_22006996 | 0.29 |
ENSDART00000116114
|
si:dkey-71b5.7
|
si:dkey-71b5.7 |
| chr25_+_35058088 | 0.29 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr19_-_9522548 | 0.28 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr17_-_24439672 | 0.28 |
ENSDART00000155020
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr7_+_22982201 | 0.26 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr23_+_40460333 | 0.26 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr21_-_30254185 | 0.26 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr8_-_39822917 | 0.26 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr24_-_37680917 | 0.26 |
ENSDART00000131342
|
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr11_+_36355348 | 0.26 |
ENSDART00000145427
|
sypl2a
|
synaptophysin-like 2a |
| chr22_+_7439186 | 0.26 |
ENSDART00000190667
|
zgc:92041
|
zgc:92041 |
| chr15_-_40267485 | 0.25 |
ENSDART00000152253
|
kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr4_-_52165969 | 0.25 |
ENSDART00000171130
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr19_-_5369486 | 0.24 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr8_+_8671229 | 0.24 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr1_-_23596391 | 0.24 |
ENSDART00000155184
|
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr1_-_35924495 | 0.24 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr5_+_26795465 | 0.23 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr24_-_26302375 | 0.23 |
ENSDART00000130696
|
cops9
|
COP9 signalosome subunit 9 |
| chr2_+_24177006 | 0.22 |
ENSDART00000132582
|
map4l
|
microtubule associated protein 4 like |
| chr3_+_19299309 | 0.22 |
ENSDART00000046297
ENSDART00000146955 |
ldlra
|
low density lipoprotein receptor a |
| chr7_-_35314347 | 0.22 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr5_+_66433287 | 0.21 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr14_+_44794936 | 0.21 |
ENSDART00000128881
|
zgc:195212
|
zgc:195212 |
| chr15_-_20949692 | 0.21 |
ENSDART00000185548
|
tbcela
|
tubulin folding cofactor E-like a |
| chr19_-_6083761 | 0.20 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr6_+_4229360 | 0.20 |
ENSDART00000191347
ENSDART00000130642 |
FO082877.1
|
|
| chr4_+_28374628 | 0.19 |
ENSDART00000076037
|
alg10
|
asparagine-linked glycosylation 10 |
| chr21_+_11401247 | 0.19 |
ENSDART00000143952
|
cel.1
|
carboxyl ester lipase, tandem duplicate 1 |
| chr6_+_39812475 | 0.19 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr16_-_17072440 | 0.19 |
ENSDART00000002493
ENSDART00000178443 |
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr6_-_40195510 | 0.19 |
ENSDART00000156156
|
col7a1
|
collagen, type VII, alpha 1 |
| chr5_+_3118231 | 0.18 |
ENSDART00000179809
|
supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr10_+_25204626 | 0.18 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr5_-_61431809 | 0.18 |
ENSDART00000082952
|
rcc1l
|
RCC1 like |
| chr11_-_30158191 | 0.18 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr2_+_22659787 | 0.18 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr13_-_1349922 | 0.17 |
ENSDART00000140970
|
si:ch73-52p7.1
|
si:ch73-52p7.1 |
| chr2_+_904362 | 0.17 |
ENSDART00000024967
|
ripk1l
|
receptor (TNFRSF)-interacting serine-threonine kinase 1, like |
| chr2_+_24177190 | 0.17 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr1_+_46948010 | 0.17 |
ENSDART00000132843
|
si:dkey-22n8.2
|
si:dkey-22n8.2 |
| chr20_-_47188966 | 0.17 |
ENSDART00000152965
|
si:dkeyp-104f11.9
|
si:dkeyp-104f11.9 |
| chr25_+_19485198 | 0.16 |
ENSDART00000156730
|
glsl
|
glutaminase like |
| chr4_+_69823638 | 0.16 |
ENSDART00000165786
|
znf1087
|
zinc finger protein 1087 |
| chr19_+_37509638 | 0.16 |
ENSDART00000139999
|
si:ch211-250g4.3
|
si:ch211-250g4.3 |
| chr22_+_17606863 | 0.15 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| chr22_+_7462997 | 0.15 |
ENSDART00000106082
|
zgc:112368
|
zgc:112368 |
| chr16_-_33001153 | 0.15 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr11_+_16153207 | 0.15 |
ENSDART00000192356
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr23_+_4226341 | 0.14 |
ENSDART00000012445
|
zgc:113278
|
zgc:113278 |
| chr16_+_40217043 | 0.14 |
ENSDART00000191128
|
BX470182.1
|
|
| chr15_-_15357178 | 0.14 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr1_-_23595779 | 0.14 |
ENSDART00000134860
ENSDART00000138852 |
lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr4_+_14957360 | 0.13 |
ENSDART00000002770
ENSDART00000111882 ENSDART00000148292 |
tspan33a
|
tetraspanin 33a |
| chr5_+_10084100 | 0.13 |
ENSDART00000109236
|
si:ch211-207k7.4
|
si:ch211-207k7.4 |
| chr12_+_47663419 | 0.13 |
ENSDART00000171932
|
HHEX
|
hematopoietically expressed homeobox |
| chr17_+_33340675 | 0.13 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr11_-_1409236 | 0.12 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr7_+_57108823 | 0.12 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr15_+_11840311 | 0.12 |
ENSDART00000167671
|
prkd2
|
protein kinase D2 |
| chr5_-_66301142 | 0.12 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr17_-_48915427 | 0.12 |
ENSDART00000054781
|
lgals8b
|
galectin 8b |
| chr9_+_35014513 | 0.12 |
ENSDART00000100701
|
gabpa
|
GA binding protein transcription factor, alpha subunit |
| chr17_-_39185336 | 0.12 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr25_+_17920361 | 0.12 |
ENSDART00000185644
|
borcs5
|
BLOC-1 related complex subunit 5 |
| chr23_-_17657348 | 0.11 |
ENSDART00000054736
|
bhlhe23
|
basic helix-loop-helix family, member e23 |
| chr18_+_30028637 | 0.11 |
ENSDART00000139750
|
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr21_-_45363871 | 0.11 |
ENSDART00000075443
ENSDART00000182078 ENSDART00000151106 |
PPP2CA
|
zgc:56064 |
| chr16_-_44878245 | 0.10 |
ENSDART00000154391
ENSDART00000154925 ENSDART00000154697 |
arhgap33
|
Rho GTPase activating protein 33 |
| chr1_-_54765262 | 0.10 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr9_+_32859967 | 0.10 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr20_-_38836161 | 0.10 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr8_-_5220125 | 0.09 |
ENSDART00000035676
|
bnip3la
|
BCL2 interacting protein 3 like a |
| chr11_+_30513656 | 0.09 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr17_+_2727807 | 0.09 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr20_-_34663209 | 0.09 |
ENSDART00000132545
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr3_-_12930217 | 0.09 |
ENSDART00000166322
|
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr23_-_6865946 | 0.09 |
ENSDART00000056426
|
ftr58
|
finTRIM family, member 58 |
| chr8_+_24861264 | 0.08 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr2_-_57344037 | 0.08 |
ENSDART00000148873
|
tcf3a
|
transcription factor 3a |
| chr19_+_26072624 | 0.08 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr6_+_54538948 | 0.08 |
ENSDART00000149270
|
tulp1b
|
tubby like protein 1b |
| chr5_+_63857055 | 0.08 |
ENSDART00000138950
|
rgs3b
|
regulator of G protein signaling 3b |
| chr1_-_36152131 | 0.07 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr13_+_35637048 | 0.07 |
ENSDART00000085037
|
thbs2a
|
thrombospondin 2a |
| chr6_-_1768724 | 0.07 |
ENSDART00000162488
ENSDART00000163613 |
zgc:158417
|
zgc:158417 |
| chr18_+_3579829 | 0.06 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr13_+_10023256 | 0.06 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr19_-_24224142 | 0.06 |
ENSDART00000136409
ENSDART00000114390 |
prf1.8
|
perforin 1.8 |
| chr15_+_17251191 | 0.06 |
ENSDART00000156587
|
si:ch73-223p23.2
|
si:ch73-223p23.2 |
| chr5_-_11905920 | 0.06 |
ENSDART00000114378
|
fbxw8
|
F-box and WD repeat domain containing 8 |
| chr25_+_19695652 | 0.06 |
ENSDART00000104340
|
mxg
|
myxovirus (influenza virus) resistance G |
| chr8_+_23784471 | 0.06 |
ENSDART00000189457
|
si:ch211-163l21.8
|
si:ch211-163l21.8 |
| chr10_-_21953643 | 0.06 |
ENSDART00000188921
ENSDART00000193569 |
FO744833.2
|
|
| chr20_+_9128829 | 0.06 |
ENSDART00000064144
ENSDART00000137450 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr3_+_22375596 | 0.06 |
ENSDART00000188243
ENSDART00000181506 |
arhgap27l
|
Rho GTPase activating protein 27, like |
| chr14_-_17576662 | 0.05 |
ENSDART00000193893
|
rnf4
|
ring finger protein 4 |
| chr25_+_14087045 | 0.05 |
ENSDART00000155770
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr8_-_50888806 | 0.05 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr7_+_10562118 | 0.05 |
ENSDART00000185188
ENSDART00000168801 |
zfand6
|
zinc finger, AN1-type domain 6 |
| chr2_-_26509057 | 0.05 |
ENSDART00000139169
ENSDART00000181777 |
best4
|
bestrophin 4 |
| chr3_+_22270463 | 0.05 |
ENSDART00000104004
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr5_-_51484156 | 0.05 |
ENSDART00000162064
|
CR388055.1
|
|
| chr10_+_17714866 | 0.05 |
ENSDART00000039969
|
slc20a1b
|
solute carrier family 20 (phosphate transporter), member 1b |
| chr22_+_3238474 | 0.04 |
ENSDART00000157954
|
si:ch1073-178p5.3
|
si:ch1073-178p5.3 |
| chr6_+_39098397 | 0.04 |
ENSDART00000003716
ENSDART00000188655 |
prss60.2
|
protease, serine, 60.2 |
| chr6_-_27108844 | 0.04 |
ENSDART00000073883
|
dtymk
|
deoxythymidylate kinase (thymidylate kinase) |
| chr6_-_50704689 | 0.04 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr14_+_8715527 | 0.04 |
ENSDART00000170754
|
kcnk4a
|
potassium channel, subfamily K, member 4a |
| chr20_-_47731768 | 0.04 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr19_-_42588510 | 0.03 |
ENSDART00000102583
|
sytl1
|
synaptotagmin-like 1 |
| chr22_-_6991514 | 0.03 |
ENSDART00000148233
|
fgfr1bl
|
fibroblast growth factor receptor 1b, like |
| chr21_-_18648861 | 0.03 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr20_+_37295006 | 0.03 |
ENSDART00000153137
|
cx23
|
connexin 23 |
| chr12_-_44010532 | 0.03 |
ENSDART00000183875
|
si:ch211-182p11.1
|
si:ch211-182p11.1 |
| chr1_-_35695614 | 0.02 |
ENSDART00000133813
ENSDART00000145291 ENSDART00000176451 |
si:dkey-27h10.2
|
si:dkey-27h10.2 |
| chr25_-_20691609 | 0.02 |
ENSDART00000186942
|
edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae) |
| chr12_-_20665164 | 0.02 |
ENSDART00000105352
|
gip
|
gastric inhibitory polypeptide |
| chr3_+_18425814 | 0.02 |
ENSDART00000144222
|
gaa
|
glucosidase, alpha; acid (Pompe disease, glycogen storage disease type II) |
| chr9_-_18568927 | 0.02 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr20_+_16703050 | 0.02 |
ENSDART00000005244
|
kcnk13b
|
potassium channel, subfamily K, member 13b |
| chr9_+_19039608 | 0.02 |
ENSDART00000055866
|
chmp2ba
|
charged multivesicular body protein 2Ba |
| chr16_-_52540056 | 0.02 |
ENSDART00000188304
|
CR293507.1
|
|
| chr16_+_23087326 | 0.02 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox11a+sox11b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.3 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.2 | 0.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 0.9 | GO:0003352 | regulation of cilium movement(GO:0003352) |
| 0.1 | 1.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.7 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.1 | 0.5 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.2 | GO:0015889 | cobalt ion transport(GO:0006824) cobalamin transport(GO:0015889) |
| 0.1 | 0.2 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.1 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.3 | GO:0051445 | regulation of meiotic cell cycle(GO:0051445) |
| 0.1 | 0.5 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.7 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.5 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 0.3 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) chronic inflammatory response(GO:0002544) regulation of granuloma formation(GO:0002631) regulation of chronic inflammatory response(GO:0002676) |
| 0.1 | 0.8 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.3 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.3 | GO:0034080 | chromatin remodeling at centromere(GO:0031055) CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.4 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.5 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.0 | 0.1 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.3 | GO:0021576 | hindbrain formation(GO:0021576) |
| 0.0 | 0.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.6 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.9 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.8 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0034381 | plasma lipoprotein particle clearance(GO:0034381) |
| 0.0 | 0.2 | GO:0006543 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0089700 | endocardial cushion formation(GO:0003272) protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0003171 | atrioventricular valve development(GO:0003171) |
| 0.0 | 0.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.6 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.6 | GO:0099500 | synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.0 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 0.0 | 0.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 1.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.5 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.3 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.1 | 1.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.2 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.9 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 1.0 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 0.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 1.3 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.8 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.6 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.1 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.1 | 0.9 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.1 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.8 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.5 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.2 | GO:0030228 | lipoprotein particle receptor activity(GO:0030228) |
| 0.0 | 1.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.6 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.8 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.1 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.6 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.3 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |