Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
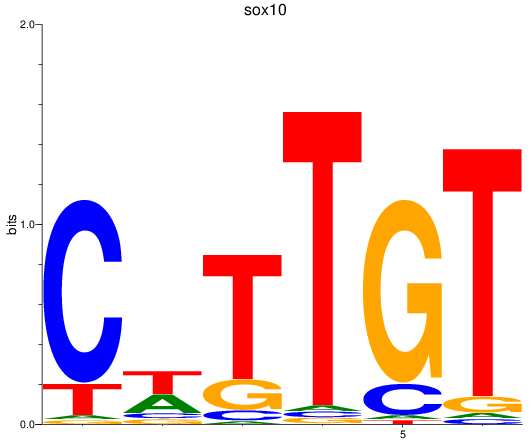
Results for sox10
Z-value: 0.64
Transcription factors associated with sox10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox10
|
ENSDARG00000077467 | SRY-box transcription factor 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox10 | dr11_v1_chr3_+_1492174_1492174 | -0.11 | 6.5e-01 | Click! |
Activity profile of sox10 motif
Sorted Z-values of sox10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_80685 | 1.01 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr23_-_35483163 | 0.94 |
ENSDART00000138660
ENSDART00000113643 ENSDART00000189269 |
fbxo25
|
F-box protein 25 |
| chr3_+_24197934 | 0.93 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr20_+_7584211 | 0.89 |
ENSDART00000132481
ENSDART00000127975 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr6_-_21091948 | 0.80 |
ENSDART00000057348
|
inha
|
inhibin, alpha |
| chr14_+_46313396 | 0.80 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr10_+_44042033 | 0.79 |
ENSDART00000190006
ENSDART00000046172 |
cryba4
|
crystallin, beta A4 |
| chr12_-_5418340 | 0.78 |
ENSDART00000028043
|
noc3l
|
NOC3-like DNA replication regulator |
| chr20_-_14665002 | 0.76 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr17_-_6599484 | 0.75 |
ENSDART00000156927
|
ANKRD66
|
si:ch211-189e2.2 |
| chr4_+_15819728 | 0.75 |
ENSDART00000101613
|
lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr20_-_54014539 | 0.74 |
ENSDART00000060466
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr14_+_46313135 | 0.74 |
ENSDART00000172902
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr14_-_28567845 | 0.71 |
ENSDART00000126095
|
insb
|
preproinsulin b |
| chr16_+_9762261 | 0.70 |
ENSDART00000020654
|
psmd4b
|
proteasome 26S subunit, non-ATPase 4b |
| chr21_-_32301109 | 0.69 |
ENSDART00000139890
|
clk4b
|
CDC-like kinase 4b |
| chr3_+_15505275 | 0.69 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr16_-_2650341 | 0.66 |
ENSDART00000128169
ENSDART00000155432 ENSDART00000103722 |
lyplal1
|
lysophospholipase-like 1 |
| chr24_+_11083146 | 0.66 |
ENSDART00000009473
|
zfand1
|
zinc finger, AN1-type domain 1 |
| chr8_+_2575993 | 0.65 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr18_+_20838786 | 0.65 |
ENSDART00000138692
|
ttc23
|
tetratricopeptide repeat domain 23 |
| chr3_-_41535647 | 0.65 |
ENSDART00000153723
ENSDART00000154198 |
si:ch211-222n22.1
|
si:ch211-222n22.1 |
| chr9_-_18911608 | 0.64 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr23_-_31506854 | 0.63 |
ENSDART00000131352
ENSDART00000138625 ENSDART00000133002 |
eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr15_-_39955785 | 0.61 |
ENSDART00000154556
|
msh5
|
mutS homolog 5 |
| chr5_+_67662430 | 0.59 |
ENSDART00000137700
ENSDART00000142586 |
si:dkey-70b23.2
|
si:dkey-70b23.2 |
| chr4_+_72742212 | 0.59 |
ENSDART00000171021
|
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr11_+_13528437 | 0.58 |
ENSDART00000011362
|
arrdc2
|
arrestin domain containing 2 |
| chr8_-_13315304 | 0.58 |
ENSDART00000142596
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr11_-_11471857 | 0.57 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr8_-_13315567 | 0.56 |
ENSDART00000132685
ENSDART00000168635 |
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr25_+_35058088 | 0.56 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr11_+_26604224 | 0.55 |
ENSDART00000030453
ENSDART00000168895 ENSDART00000159505 |
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr15_+_44053244 | 0.54 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr7_-_35126374 | 0.53 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr21_+_19347655 | 0.52 |
ENSDART00000093155
|
hpse
|
heparanase |
| chr16_-_26731928 | 0.52 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr21_-_25756119 | 0.51 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr3_+_24458204 | 0.51 |
ENSDART00000155028
ENSDART00000153551 |
cbx6b
|
chromobox homolog 6b |
| chr23_-_30727596 | 0.51 |
ENSDART00000060193
|
thap3
|
THAP domain containing, apoptosis associated protein 3 |
| chr5_+_43807003 | 0.50 |
ENSDART00000097625
|
zgc:158640
|
zgc:158640 |
| chr5_-_6377865 | 0.50 |
ENSDART00000031775
|
zgc:73226
|
zgc:73226 |
| chr3_-_16250527 | 0.50 |
ENSDART00000146699
ENSDART00000141181 |
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr5_-_71705191 | 0.49 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr7_+_22688781 | 0.48 |
ENSDART00000173509
|
ugt5g1
|
UDP glucuronosyltransferase 5 family, polypeptide G1 |
| chr22_-_15587360 | 0.48 |
ENSDART00000142717
ENSDART00000138978 |
tpm4a
|
tropomyosin 4a |
| chr12_-_30841679 | 0.48 |
ENSDART00000105594
|
crygmx
|
crystallin, gamma MX |
| chr14_-_33348221 | 0.47 |
ENSDART00000187749
|
rpl39
|
ribosomal protein L39 |
| chr4_-_193762 | 0.47 |
ENSDART00000169667
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr2_+_3472832 | 0.47 |
ENSDART00000115278
|
cx47.1
|
connexin 47.1 |
| chr21_+_7131970 | 0.47 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr12_-_42368296 | 0.47 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr24_+_5840258 | 0.46 |
ENSDART00000087034
|
trpc1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr23_+_36340520 | 0.46 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr25_+_20716176 | 0.46 |
ENSDART00000073651
|
ergic2
|
ERGIC and golgi 2 |
| chr5_+_69868911 | 0.46 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr5_-_51148298 | 0.46 |
ENSDART00000042420
|
ccdc180
|
coiled-coil domain containing 180 |
| chr5_+_9134187 | 0.45 |
ENSDART00000132240
|
pimr123
|
Pim proto-oncogene, serine/threonine kinase, related 123 |
| chr15_-_108414 | 0.45 |
ENSDART00000170044
|
apoa1b
|
apolipoprotein A-Ib |
| chr20_+_39457598 | 0.45 |
ENSDART00000140931
ENSDART00000156176 |
pimr128
|
Pim proto-oncogene, serine/threonine kinase, related 128 |
| chr5_-_41645058 | 0.45 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr16_+_40560622 | 0.45 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr9_-_29497916 | 0.45 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr6_-_6993046 | 0.45 |
ENSDART00000053304
|
si:ch211-114n24.6
|
si:ch211-114n24.6 |
| chr13_-_1151410 | 0.45 |
ENSDART00000007231
|
psmb1
|
proteasome subunit beta 1 |
| chr17_-_122680 | 0.44 |
ENSDART00000066430
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr19_+_18903533 | 0.44 |
ENSDART00000157523
ENSDART00000166562 |
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr1_+_41849152 | 0.44 |
ENSDART00000053685
|
smox
|
spermine oxidase |
| chr23_-_30787932 | 0.44 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr11_+_24820542 | 0.43 |
ENSDART00000135443
|
kdm5ba
|
lysine (K)-specific demethylase 5Ba |
| chr23_+_22658700 | 0.43 |
ENSDART00000192248
|
eno1a
|
enolase 1a, (alpha) |
| chr19_+_63567 | 0.42 |
ENSDART00000165657
ENSDART00000165183 |
zhx2b
|
zinc fingers and homeoboxes 2b |
| chr8_-_14067517 | 0.42 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr12_-_34035364 | 0.42 |
ENSDART00000087065
|
timp2a
|
TIMP metallopeptidase inhibitor 2a |
| chr6_-_51101834 | 0.42 |
ENSDART00000092493
|
ptprt
|
protein tyrosine phosphatase, receptor type, t |
| chr23_-_14830627 | 0.42 |
ENSDART00000134659
|
sla2
|
Src-like-adaptor 2 |
| chr19_+_15521997 | 0.41 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr12_-_15567104 | 0.41 |
ENSDART00000053003
|
hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr5_-_64883082 | 0.40 |
ENSDART00000064983
ENSDART00000139066 |
krt1-c5
|
keratin, type 1, gene c5 |
| chr24_+_18714212 | 0.39 |
ENSDART00000171181
|
cspp1a
|
centrosome and spindle pole associated protein 1a |
| chr19_+_26714949 | 0.39 |
ENSDART00000188540
|
zgc:100906
|
zgc:100906 |
| chr25_+_16689633 | 0.39 |
ENSDART00000073416
|
ada2a
|
adenosine deaminase 2a |
| chr15_+_28368644 | 0.39 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr7_+_24814866 | 0.39 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr21_-_18972206 | 0.39 |
ENSDART00000146743
|
pimr72
|
Pim proto-oncogene, serine/threonine kinase, related 72 |
| chr14_-_33360344 | 0.39 |
ENSDART00000181291
|
pimr117
|
Pim proto-oncogene, serine/threonine kinase, related 117 |
| chr23_+_12545114 | 0.38 |
ENSDART00000105283
ENSDART00000166990 |
si:zfos-452g4.1
|
si:zfos-452g4.1 |
| chr24_-_24114217 | 0.38 |
ENSDART00000160578
|
zgc:112982
|
zgc:112982 |
| chr14_-_4170654 | 0.37 |
ENSDART00000188347
|
si:dkey-185e18.7
|
si:dkey-185e18.7 |
| chr22_+_1911269 | 0.37 |
ENSDART00000164158
ENSDART00000168205 |
znf1156
|
zinc finger protein 1156 |
| chr25_+_14400704 | 0.37 |
ENSDART00000067235
|
ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr15_-_16183583 | 0.37 |
ENSDART00000062335
|
glod4
|
glyoxalase domain containing 4 |
| chr18_-_4957022 | 0.36 |
ENSDART00000101686
|
zbbx
|
zinc finger, B-box domain containing |
| chr8_-_50147948 | 0.36 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr6_+_33885828 | 0.35 |
ENSDART00000179994
|
gpbp1l1
|
GC-rich promoter binding protein 1-like 1 |
| chr7_-_6459481 | 0.35 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr9_-_24985511 | 0.35 |
ENSDART00000141957
|
dnah7
|
dynein, axonemal, heavy chain 7 |
| chr20_+_25586099 | 0.35 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr5_+_9112291 | 0.35 |
ENSDART00000135177
|
AL845369.1
|
|
| chr3_-_53114299 | 0.35 |
ENSDART00000109390
|
AL954361.1
|
|
| chr2_-_30912922 | 0.34 |
ENSDART00000141669
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr2_-_30912307 | 0.34 |
ENSDART00000188653
|
myl12.2
|
myosin, light chain 12, genome duplicate 2 |
| chr21_-_35853245 | 0.34 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr3_-_18030938 | 0.34 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr11_+_35364445 | 0.33 |
ENSDART00000125766
|
camkvb
|
CaM kinase-like vesicle-associated b |
| chr7_-_22981796 | 0.33 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr15_-_28200049 | 0.33 |
ENSDART00000004200
|
sarm1
|
sterile alpha and TIR motif containing 1 |
| chr15_+_3825117 | 0.33 |
ENSDART00000183315
|
CABZ01061591.1
|
|
| chr7_+_49664174 | 0.33 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr16_+_53125918 | 0.33 |
ENSDART00000102170
|
CABZ01053976.1
|
|
| chr1_-_10847753 | 0.33 |
ENSDART00000141052
|
dmd
|
dystrophin |
| chr10_+_16501699 | 0.32 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr12_-_36045283 | 0.32 |
ENSDART00000160646
|
gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr14_+_9287683 | 0.31 |
ENSDART00000122485
|
msnb
|
moesin b |
| chr4_+_17280868 | 0.31 |
ENSDART00000145349
|
bcat1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr22_-_10570749 | 0.31 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr21_-_28920245 | 0.31 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr2_+_19522082 | 0.31 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr12_+_35654749 | 0.31 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr2_+_13721888 | 0.31 |
ENSDART00000089618
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr2_-_42960353 | 0.31 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr2_-_32768951 | 0.31 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr7_+_73819078 | 0.31 |
ENSDART00000169756
|
FP236812.1
|
Histone H2B 1/2 |
| chr21_-_2310064 | 0.30 |
ENSDART00000169520
|
si:ch211-241b2.1
|
si:ch211-241b2.1 |
| chr2_-_45510699 | 0.30 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr19_-_3876877 | 0.30 |
ENSDART00000163711
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr20_+_18225329 | 0.30 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr24_-_2843107 | 0.29 |
ENSDART00000165290
|
cyb5a
|
cytochrome b5 type A (microsomal) |
| chr20_+_25568694 | 0.29 |
ENSDART00000063107
ENSDART00000063128 |
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr11_-_41220794 | 0.29 |
ENSDART00000192895
|
mrps16
|
mitochondrial ribosomal protein S16 |
| chr24_-_26369185 | 0.29 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr12_-_35883814 | 0.29 |
ENSDART00000177986
ENSDART00000129888 |
cep131
|
centrosomal protein 131 |
| chr16_-_45917322 | 0.29 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr17_-_20236228 | 0.29 |
ENSDART00000136490
ENSDART00000029380 |
bnip4
|
BCL2 interacting protein 4 |
| chr25_-_12788370 | 0.28 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr1_+_17376922 | 0.28 |
ENSDART00000145068
|
fat1a
|
FAT atypical cadherin 1a |
| chr1_-_19845378 | 0.28 |
ENSDART00000139314
ENSDART00000132958 ENSDART00000147502 |
grhprb
|
glyoxylate reductase/hydroxypyruvate reductase b |
| chr2_+_19633493 | 0.28 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr6_-_9922266 | 0.28 |
ENSDART00000151549
|
pimr73
|
Pim proto-oncogene, serine/threonine kinase, related 73 |
| chr4_-_76154252 | 0.28 |
ENSDART00000161269
|
BX957252.1
|
|
| chr3_-_24458281 | 0.28 |
ENSDART00000153993
|
baiap2l2a
|
BAI1-associated protein 2-like 2a |
| chr6_-_7020162 | 0.28 |
ENSDART00000148982
|
bin1b
|
bridging integrator 1b |
| chr22_-_16180849 | 0.28 |
ENSDART00000090390
|
vcam1b
|
vascular cell adhesion molecule 1b |
| chr8_+_42998944 | 0.28 |
ENSDART00000048819
|
rassf2a
|
Ras association (RalGDS/AF-6) domain family member 2a |
| chr4_-_11403811 | 0.27 |
ENSDART00000067272
ENSDART00000140018 |
pimr173
|
Pim proto-oncogene, serine/threonine kinase, related 173 |
| chr13_+_28854438 | 0.27 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr6_+_45932276 | 0.27 |
ENSDART00000103491
|
rbp7b
|
retinol binding protein 7b, cellular |
| chr17_-_24564674 | 0.27 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr3_-_31069776 | 0.27 |
ENSDART00000167462
|
elob
|
elongin B |
| chr22_+_14051894 | 0.27 |
ENSDART00000142548
|
aox6
|
aldehyde oxidase 6 |
| chr9_-_27857265 | 0.26 |
ENSDART00000177513
ENSDART00000143166 ENSDART00000115313 |
si:rp71-45g20.4
|
si:rp71-45g20.4 |
| chr9_+_38737924 | 0.26 |
ENSDART00000147652
|
kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr6_+_14980761 | 0.26 |
ENSDART00000087782
|
mrps9
|
mitochondrial ribosomal protein S9 |
| chr11_+_6882362 | 0.26 |
ENSDART00000144181
|
klhl26
|
kelch-like family member 26 |
| chr22_+_29067388 | 0.26 |
ENSDART00000133673
|
pimr100
|
Pim proto-oncogene, serine/threonine kinase, related 100 |
| chr25_-_13490744 | 0.25 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr2_+_13722116 | 0.25 |
ENSDART00000155015
|
zbtb41
|
zinc finger and BTB domain containing 41 |
| chr11_+_6882049 | 0.25 |
ENSDART00000075998
|
klhl26
|
kelch-like family member 26 |
| chr12_-_7655322 | 0.25 |
ENSDART00000181162
|
ank3b
|
ankyrin 3b |
| chr2_-_25140022 | 0.24 |
ENSDART00000134543
|
nceh1a
|
neutral cholesterol ester hydrolase 1a |
| chr23_+_36144487 | 0.24 |
ENSDART00000082473
|
hoxc3a
|
homeobox C3a |
| chr10_-_42131408 | 0.24 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr7_-_60096318 | 0.24 |
ENSDART00000189125
|
BX511067.1
|
|
| chr6_-_23931442 | 0.24 |
ENSDART00000160547
|
sec16b
|
SEC16 homolog B, endoplasmic reticulum export factor |
| chr25_+_35132090 | 0.24 |
ENSDART00000154377
|
si:dkey-261m9.6
|
si:dkey-261m9.6 |
| chr5_-_30984010 | 0.24 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr21_-_17603182 | 0.24 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr17_+_26208630 | 0.23 |
ENSDART00000087084
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr9_+_27411502 | 0.23 |
ENSDART00000143994
|
si:dkey-193n17.9
|
si:dkey-193n17.9 |
| chr14_-_26400501 | 0.23 |
ENSDART00000054189
|
pimr212
|
Pim proto-oncogene, serine/threonine kinase, related 212 |
| chr11_+_2600612 | 0.23 |
ENSDART00000173442
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr8_-_16697912 | 0.23 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr21_-_13972745 | 0.23 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr16_-_32233463 | 0.23 |
ENSDART00000102016
|
calhm6
|
calcium homeostasis modulator family member 6 |
| chr16_-_50203058 | 0.23 |
ENSDART00000154570
|
vsig10l
|
V-set and immunoglobulin domain containing 10 like |
| chr3_+_3545825 | 0.23 |
ENSDART00000109060
|
CR589947.1
|
|
| chr2_-_19576640 | 0.23 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr3_-_58226583 | 0.22 |
ENSDART00000187429
|
im:6904045
|
im:6904045 |
| chr7_-_34329527 | 0.22 |
ENSDART00000173454
|
madd
|
MAP-kinase activating death domain |
| chr18_-_14836862 | 0.22 |
ENSDART00000124843
|
mtss1la
|
metastasis suppressor 1-like a |
| chr2_-_45510223 | 0.22 |
ENSDART00000113058
|
gpsm2
|
G protein signaling modulator 2 |
| chr5_+_58397646 | 0.22 |
ENSDART00000180759
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr1_-_51474974 | 0.22 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr13_+_36958086 | 0.21 |
ENSDART00000024386
|
frmd6
|
FERM domain containing 6 |
| chr7_+_20019125 | 0.21 |
ENSDART00000186391
|
bcl6b
|
B-cell CLL/lymphoma 6, member B |
| chr11_-_7147540 | 0.21 |
ENSDART00000143942
|
bmp7a
|
bone morphogenetic protein 7a |
| chr4_-_71110826 | 0.21 |
ENSDART00000167431
|
si:dkeyp-80d11.1
|
si:dkeyp-80d11.1 |
| chr20_-_284865 | 0.21 |
ENSDART00000104806
|
wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr5_+_57658898 | 0.21 |
ENSDART00000074268
ENSDART00000124568 |
zgc:153929
|
zgc:153929 |
| chr5_+_36870737 | 0.20 |
ENSDART00000145182
|
slc8a2a
|
solute carrier family 8 (sodium/calcium exchanger), member 2a |
| chr18_-_37241080 | 0.20 |
ENSDART00000126421
ENSDART00000078064 |
six9
|
SIX homeobox 9 |
| chr11_+_583142 | 0.20 |
ENSDART00000168157
|
mkrn2os.2
|
MKRN2 opposite strand, tandem duplicate 2 |
| chr23_-_21471022 | 0.20 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr12_+_26706745 | 0.20 |
ENSDART00000141401
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr11_-_3381343 | 0.20 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr2_+_35595454 | 0.20 |
ENSDART00000098734
|
cacybp
|
calcyclin binding protein |
| chr3_+_30500968 | 0.20 |
ENSDART00000103447
|
si:dkey-13n23.3
|
si:dkey-13n23.3 |
| chr22_+_1028724 | 0.20 |
ENSDART00000149625
|
si:ch73-352p18.4
|
si:ch73-352p18.4 |
| chr22_+_1462177 | 0.20 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr9_+_35876927 | 0.20 |
ENSDART00000138834
|
mab21l3
|
mab-21-like 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sox10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 0.9 | GO:1902038 | positive regulation of hematopoietic stem cell differentiation(GO:1902038) |
| 0.1 | 0.5 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.8 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 0.3 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.1 | 0.4 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.1 | 0.5 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.6 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 0.5 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.6 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.4 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.2 | GO:1901827 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.6 | GO:0000710 | meiotic mismatch repair(GO:0000710) |
| 0.1 | 0.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.4 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0046125 | deoxyribonucleoside metabolic process(GO:0009120) thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.1 | GO:0072196 | proximal/distal pattern formation involved in nephron development(GO:0072047) proximal/distal pattern formation involved in pronephric nephron development(GO:0072196) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0061178 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061178) |
| 0.0 | 0.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.2 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.1 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.2 | GO:1902765 | L-arginine import(GO:0043091) amino acid import across plasma membrane(GO:0089718) arginine import(GO:0090467) L-arginine import across plasma membrane(GO:0097638) L-arginine transport(GO:1902023) L-arginine import into cell(GO:1902765) amino acid import into cell(GO:1902837) L-arginine transmembrane transport(GO:1903400) arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 3.4 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 0.5 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.0 | 0.3 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0003151 | outflow tract morphogenesis(GO:0003151) |
| 0.0 | 0.1 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) |
| 0.0 | 0.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 1.0 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.0 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.6 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.2 | GO:0098719 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.0 | 0.6 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.4 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0003261 | cardiac muscle progenitor cell migration to the midline involved in heart field formation(GO:0003261) |
| 0.0 | 0.2 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.0 | 0.1 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.5 | GO:0015908 | fatty acid transport(GO:0015908) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.3 | GO:0071910 | determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.4 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.1 | GO:0051703 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.2 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.3 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.1 | GO:0030431 | sleep(GO:0030431) |
| 0.0 | 0.2 | GO:1990089 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.2 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 3.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.2 | GO:0098659 | inorganic cation import into cell(GO:0098659) calcium ion import across plasma membrane(GO:0098703) inorganic ion import into cell(GO:0099587) calcium ion import into cell(GO:1990035) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.1 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.1 | 0.9 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.7 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.3 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 1.0 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.5 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.6 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.5 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.5 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0097268 | cytoophidium(GO:0097268) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 1.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.4 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.1 | 0.5 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.1 | 0.3 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.1 | 0.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.1 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.2 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 0.4 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 2.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.7 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0005252 | open rectifier potassium channel activity(GO:0005252) |
| 0.0 | 0.2 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0017113 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.5 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.0 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.6 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor-activated receptor activity(GO:0005017) platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.0 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.4 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0051018 | protein kinase A binding(GO:0051018) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.2 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |