Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
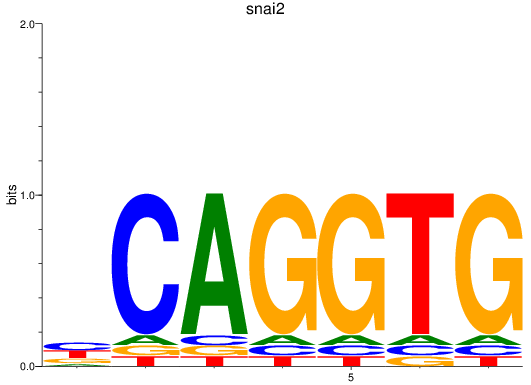
Results for snai2
Z-value: 0.41
Transcription factors associated with snai2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai2
|
ENSDARG00000040046 | snail family zinc finger 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai2 | dr11_v1_chr24_+_35387517_35387517 | 0.85 | 7.2e-06 | Click! |
Activity profile of snai2 motif
Sorted Z-values of snai2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_1052591 | 1.27 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr2_+_26237322 | 0.92 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr14_+_15155684 | 0.91 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr20_-_3238110 | 0.91 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr15_+_20239141 | 0.90 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr10_+_6340478 | 0.86 |
ENSDART00000163182
ENSDART00000163722 |
tpm2
|
tropomyosin 2 (beta) |
| chr9_-_28255029 | 0.85 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr12_-_4243268 | 0.82 |
ENSDART00000131275
|
zgc:92313
|
zgc:92313 |
| chr23_+_2728095 | 0.79 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr6_-_37749711 | 0.78 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr19_+_791538 | 0.75 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr18_+_45666489 | 0.74 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr23_+_27778670 | 0.74 |
ENSDART00000053863
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr7_-_33868903 | 0.73 |
ENSDART00000173500
ENSDART00000178746 |
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr21_-_21178410 | 0.73 |
ENSDART00000185277
ENSDART00000141341 ENSDART00000145872 ENSDART00000079678 |
ftsj1
|
FtsJ RNA methyltransferase homolog 1 |
| chr20_-_23253630 | 0.72 |
ENSDART00000103365
|
ociad1
|
OCIA domain containing 1 |
| chr6_+_1724889 | 0.69 |
ENSDART00000157415
|
acvr2ab
|
activin A receptor type 2Ab |
| chr20_-_41992878 | 0.68 |
ENSDART00000100967
|
si:dkeyp-114g9.1
|
si:dkeyp-114g9.1 |
| chr14_+_30285613 | 0.68 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr5_+_6670945 | 0.67 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr13_-_36525982 | 0.66 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr3_+_43086548 | 0.66 |
ENSDART00000163579
|
si:dkey-43p13.5
|
si:dkey-43p13.5 |
| chr17_-_24575893 | 0.66 |
ENSDART00000141914
|
aftphb
|
aftiphilin b |
| chr20_+_21268795 | 0.64 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr8_-_38022298 | 0.63 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr7_-_51773166 | 0.63 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| chr5_+_22974019 | 0.63 |
ENSDART00000147157
ENSDART00000020434 |
brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr7_-_53117131 | 0.62 |
ENSDART00000169211
ENSDART00000168890 ENSDART00000172179 ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr21_-_35325466 | 0.62 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr23_-_44574059 | 0.62 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr19_-_18313303 | 0.62 |
ENSDART00000164644
ENSDART00000167480 ENSDART00000163104 |
si:dkey-208k4.2
|
si:dkey-208k4.2 |
| chr23_+_27779452 | 0.61 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr5_+_36654817 | 0.59 |
ENSDART00000131339
|
capns1a
|
calpain, small subunit 1 a |
| chr15_+_784149 | 0.57 |
ENSDART00000155114
|
znf970
|
zinc finger protein 970 |
| chr1_-_24349759 | 0.57 |
ENSDART00000142740
ENSDART00000177989 |
lrba
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr3_-_30888415 | 0.56 |
ENSDART00000124458
|
kmt5c
|
lysine methyltransferase 5C |
| chr21_-_43666420 | 0.54 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr8_-_1698155 | 0.53 |
ENSDART00000186159
|
CABZ01065417.1
|
|
| chr4_+_26053044 | 0.53 |
ENSDART00000039877
|
SCYL2
|
si:ch211-244b2.1 |
| chr11_-_43473824 | 0.52 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr2_-_15040345 | 0.51 |
ENSDART00000109657
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr3_-_32362872 | 0.51 |
ENSDART00000035545
ENSDART00000012630 |
prmt1
|
protein arginine methyltransferase 1 |
| chr9_+_45428041 | 0.50 |
ENSDART00000193087
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr16_+_41060161 | 0.50 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr14_-_21618005 | 0.49 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr17_+_1360192 | 0.48 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr23_+_39963599 | 0.48 |
ENSDART00000166539
|
fyco1a
|
FYVE and coiled-coil domain containing 1a |
| chr19_-_15229421 | 0.48 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr2_-_58075414 | 0.48 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr13_+_34689663 | 0.47 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr14_+_989733 | 0.47 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr5_-_54712159 | 0.47 |
ENSDART00000149207
|
ccnb1
|
cyclin B1 |
| chr1_-_59313465 | 0.47 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr3_-_34528306 | 0.47 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr7_+_30988570 | 0.46 |
ENSDART00000180613
ENSDART00000185625 |
tjp1a
|
tight junction protein 1a |
| chr22_-_12304591 | 0.46 |
ENSDART00000136408
|
zranb3
|
zinc finger, RAN-binding domain containing 3 |
| chr1_+_53321878 | 0.45 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr9_-_28990649 | 0.45 |
ENSDART00000078823
|
ptpn4a
|
protein tyrosine phosphatase, non-receptor type 4a |
| chr5_+_44655148 | 0.45 |
ENSDART00000124059
|
dapk1
|
death-associated protein kinase 1 |
| chr6_+_58289335 | 0.45 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr13_-_4134141 | 0.44 |
ENSDART00000132354
|
tjap1
|
tight junction associated protein 1 (peripheral) |
| chr9_+_45493341 | 0.44 |
ENSDART00000145616
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr18_+_6641542 | 0.44 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr5_-_57204352 | 0.44 |
ENSDART00000171252
ENSDART00000180727 |
man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr24_-_31904924 | 0.43 |
ENSDART00000156060
ENSDART00000129741 ENSDART00000154276 |
si:ch73-78o10.1
|
si:ch73-78o10.1 |
| chr20_+_54336137 | 0.43 |
ENSDART00000113792
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr20_-_43743700 | 0.42 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr7_+_24528430 | 0.40 |
ENSDART00000133022
|
si:dkeyp-75h12.2
|
si:dkeyp-75h12.2 |
| chr10_+_37173029 | 0.40 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr15_-_19128705 | 0.39 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr13_+_7578111 | 0.38 |
ENSDART00000175431
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr23_+_26026383 | 0.38 |
ENSDART00000141553
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr12_-_7234915 | 0.38 |
ENSDART00000048866
|
ipmkb
|
inositol polyphosphate multikinase b |
| chr1_-_40341306 | 0.37 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr11_-_11791718 | 0.37 |
ENSDART00000180476
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr10_+_5954787 | 0.36 |
ENSDART00000161887
ENSDART00000160345 ENSDART00000190046 |
map3k1
|
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr16_-_31791165 | 0.36 |
ENSDART00000148389
|
chd4b
|
chromodomain helicase DNA binding protein 4b |
| chr18_-_26781616 | 0.36 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr23_-_32156278 | 0.35 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_+_19596214 | 0.35 |
ENSDART00000183110
|
CABZ01059626.1
|
|
| chr14_-_15155384 | 0.35 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr13_-_36050303 | 0.35 |
ENSDART00000134955
ENSDART00000139087 |
lgmn
|
legumain |
| chr1_+_46493944 | 0.34 |
ENSDART00000114083
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr17_-_25331439 | 0.34 |
ENSDART00000155422
ENSDART00000082324 |
zpcx
|
zona pellucida protein C |
| chr13_+_2448251 | 0.34 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr6_-_55399214 | 0.34 |
ENSDART00000168367
|
ctsa
|
cathepsin A |
| chr24_+_36018164 | 0.33 |
ENSDART00000182815
ENSDART00000126941 |
gnal2
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type 2 |
| chr18_-_38271298 | 0.33 |
ENSDART00000143016
|
caprin1b
|
cell cycle associated protein 1b |
| chr5_+_18014931 | 0.33 |
ENSDART00000142562
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr21_-_4250682 | 0.33 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr6_+_12482599 | 0.33 |
ENSDART00000090316
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr4_-_12978925 | 0.32 |
ENSDART00000013839
|
tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr17_+_50655619 | 0.32 |
ENSDART00000184917
ENSDART00000167638 |
ddhd1a
|
DDHD domain containing 1a |
| chr1_+_10019653 | 0.31 |
ENSDART00000190227
|
trim2b
|
tripartite motif containing 2b |
| chr2_+_23677179 | 0.31 |
ENSDART00000153918
|
oxsr1a
|
oxidative stress responsive 1a |
| chr3_+_1179601 | 0.31 |
ENSDART00000173378
|
triobpb
|
TRIO and F-actin binding protein b |
| chr1_+_24469313 | 0.31 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr22_+_30195257 | 0.31 |
ENSDART00000027803
ENSDART00000172496 |
add3a
|
adducin 3 (gamma) a |
| chr4_-_17263210 | 0.31 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr15_+_38221038 | 0.31 |
ENSDART00000188149
|
stim1a
|
stromal interaction molecule 1a |
| chr1_-_7673376 | 0.30 |
ENSDART00000013264
|
arglu1b
|
arginine and glutamate rich 1b |
| chr16_-_19568795 | 0.30 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr20_+_36806398 | 0.29 |
ENSDART00000153317
|
abracl
|
ABRA C-terminal like |
| chr11_+_6902946 | 0.29 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr11_-_25853212 | 0.28 |
ENSDART00000145655
|
tmem51b
|
transmembrane protein 51b |
| chr6_-_10728921 | 0.28 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr17_-_49412313 | 0.28 |
ENSDART00000152100
|
mthfd1b
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1b |
| chr17_+_24590177 | 0.28 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr3_+_15773991 | 0.28 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr17_-_11151655 | 0.28 |
ENSDART00000156383
|
CU179699.1
|
|
| chr18_+_6857071 | 0.27 |
ENSDART00000018735
ENSDART00000181969 |
dnaja2l
|
DnaJ (Hsp40) homolog, subfamily A, member 2, like |
| chr14_-_26482096 | 0.26 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr18_+_13077800 | 0.26 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr21_-_217589 | 0.26 |
ENSDART00000185017
|
CZQB01146713.1
|
|
| chr5_-_6561376 | 0.26 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr5_-_33959868 | 0.26 |
ENSDART00000143652
|
zgc:63972
|
zgc:63972 |
| chr3_+_40255408 | 0.25 |
ENSDART00000074746
|
smcr8a
|
Smith-Magenis syndrome chromosome region, candidate 8a |
| chr23_+_14771979 | 0.25 |
ENSDART00000137410
|
exosc10
|
exosome component 10 |
| chr15_-_47193564 | 0.25 |
ENSDART00000172453
|
LSAMP
|
limbic system-associated membrane protein |
| chr16_-_33001153 | 0.25 |
ENSDART00000147941
|
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr22_-_18546649 | 0.25 |
ENSDART00000171277
|
cirbpb
|
cold inducible RNA binding protein b |
| chr23_+_2560005 | 0.25 |
ENSDART00000186906
|
GGT7
|
gamma-glutamyltransferase 7 |
| chr16_+_52343905 | 0.24 |
ENSDART00000131051
|
ifnlr1
|
interferon lambda receptor 1 |
| chr15_+_38221196 | 0.24 |
ENSDART00000122134
ENSDART00000190099 |
stim1a
|
stromal interaction molecule 1a |
| chr21_-_21515466 | 0.24 |
ENSDART00000147593
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr15_-_37543839 | 0.23 |
ENSDART00000148870
|
kmt2bb
|
lysine (K)-specific methyltransferase 2Bb |
| chr22_-_16758438 | 0.23 |
ENSDART00000132829
|
patj
|
PATJ, crumbs cell polarity complex component |
| chr20_+_32756763 | 0.23 |
ENSDART00000023006
|
fam84a
|
family with sequence similarity 84, member A |
| chr7_+_20917966 | 0.23 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr4_-_3064101 | 0.23 |
ENSDART00000135701
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr22_+_22438783 | 0.23 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr17_-_25330775 | 0.23 |
ENSDART00000154048
|
zpcx
|
zona pellucida protein C |
| chr14_-_47849216 | 0.23 |
ENSDART00000192796
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr21_+_15733880 | 0.22 |
ENSDART00000149371
ENSDART00000184111 |
idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr16_+_33163858 | 0.22 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr5_-_32505109 | 0.21 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr7_-_19369002 | 0.21 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr17_+_50656177 | 0.20 |
ENSDART00000127374
|
ddhd1a
|
DDHD domain containing 1a |
| chr22_-_14255659 | 0.20 |
ENSDART00000167088
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr16_+_26747766 | 0.20 |
ENSDART00000183257
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr5_-_12031174 | 0.20 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr22_-_3595439 | 0.20 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr3_-_45777226 | 0.19 |
ENSDART00000192849
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr17_+_25332711 | 0.19 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr5_+_30596822 | 0.18 |
ENSDART00000188375
|
hinfp
|
histone H4 transcription factor |
| chr5_+_50913357 | 0.18 |
ENSDART00000092938
|
col4a3bpa
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein a |
| chr21_-_2287589 | 0.18 |
ENSDART00000161554
|
si:ch73-299h12.4
|
si:ch73-299h12.4 |
| chr7_+_74141297 | 0.18 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr3_-_42016693 | 0.18 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr3_+_7040363 | 0.18 |
ENSDART00000157805
|
BX000701.2
|
|
| chr5_+_42259002 | 0.18 |
ENSDART00000083778
|
eral1
|
Era-like 12S mitochondrial rRNA chaperone 1 |
| chr10_-_108952 | 0.18 |
ENSDART00000127228
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr25_-_22889519 | 0.18 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr1_-_55196103 | 0.18 |
ENSDART00000140153
|
mri1
|
methylthioribose-1-phosphate isomerase 1 |
| chr20_-_33556983 | 0.17 |
ENSDART00000168798
|
rock2bl
|
rho-associated, coiled-coil containing protein kinase 2b, like |
| chr8_-_4596662 | 0.17 |
ENSDART00000138199
|
sept5a
|
septin 5a |
| chr20_-_52902693 | 0.17 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr8_+_247163 | 0.17 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr20_-_33583779 | 0.17 |
ENSDART00000097823
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr6_-_10728057 | 0.17 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr14_+_34514336 | 0.16 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr7_-_22632518 | 0.16 |
ENSDART00000161046
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr17_+_50657509 | 0.16 |
ENSDART00000179957
|
ddhd1a
|
DDHD domain containing 1a |
| chr18_+_16715864 | 0.16 |
ENSDART00000079758
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr18_+_5308392 | 0.16 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr15_-_18574716 | 0.16 |
ENSDART00000142010
ENSDART00000019006 |
ncam1b
|
neural cell adhesion molecule 1b |
| chr3_-_32965848 | 0.16 |
ENSDART00000050930
|
casp6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr14_+_48862987 | 0.15 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr7_+_38090515 | 0.15 |
ENSDART00000131387
|
cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chr2_-_23677422 | 0.15 |
ENSDART00000079131
|
cdyl
|
chromodomain protein, Y-like |
| chr5_-_64355227 | 0.15 |
ENSDART00000170787
|
fam78aa
|
family with sequence similarity 78, member Aa |
| chr4_-_74109792 | 0.15 |
ENSDART00000174320
|
RAB21
|
zgc:154045 |
| chr8_+_8196087 | 0.15 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr24_-_17400472 | 0.14 |
ENSDART00000024691
|
cul1b
|
cullin 1b |
| chr12_+_48784731 | 0.14 |
ENSDART00000158348
|
zmiz1b
|
zinc finger, MIZ-type containing 1b |
| chr7_-_22632690 | 0.14 |
ENSDART00000165245
|
si:dkey-112a7.4
|
si:dkey-112a7.4 |
| chr17_-_2596125 | 0.14 |
ENSDART00000175740
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr5_-_68244564 | 0.14 |
ENSDART00000169350
|
CABZ01083944.1
|
|
| chr25_-_3623847 | 0.14 |
ENSDART00000172586
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr6_+_49028874 | 0.14 |
ENSDART00000175254
|
slc16a1a
|
solute carrier family 16 (monocarboxylate transporter), member 1a |
| chr3_-_1317290 | 0.13 |
ENSDART00000047094
|
LO018552.1
|
|
| chr13_+_2894536 | 0.13 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr5_-_38384289 | 0.13 |
ENSDART00000135260
|
mink1
|
misshapen-like kinase 1 |
| chr4_-_77557279 | 0.13 |
ENSDART00000180113
|
AL935186.10
|
|
| chr23_-_27607039 | 0.13 |
ENSDART00000183639
|
phf8
|
PHD finger protein 8 |
| chr9_-_44905867 | 0.12 |
ENSDART00000138316
ENSDART00000131252 ENSDART00000179383 ENSDART00000159337 |
zgc:66484
|
zgc:66484 |
| chr2_+_56012016 | 0.12 |
ENSDART00000146160
ENSDART00000188702 |
loxl5b
|
lysyl oxidase-like 5b |
| chr17_+_9308425 | 0.12 |
ENSDART00000188283
ENSDART00000183311 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr8_+_53452681 | 0.12 |
ENSDART00000166705
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr15_-_434503 | 0.12 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr3_-_11008532 | 0.12 |
ENSDART00000165086
|
CR382337.3
|
|
| chr14_+_21140612 | 0.12 |
ENSDART00000168539
|
zgc:136929
|
zgc:136929 |
| chr24_+_19578935 | 0.12 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr7_+_42328578 | 0.12 |
ENSDART00000149082
|
phkb
|
phosphorylase kinase, beta |
| chr22_-_22231720 | 0.12 |
ENSDART00000160165
|
ap3d1
|
adaptor-related protein complex 3, delta 1 subunit |
| chr16_+_5597600 | 0.12 |
ENSDART00000017307
|
zgc:91890
|
zgc:91890 |
| chr9_+_48007081 | 0.12 |
ENSDART00000060593
ENSDART00000099835 |
zgc:92380
|
zgc:92380 |
| chr5_+_31049742 | 0.11 |
ENSDART00000173097
|
zzef1
|
zinc finger, ZZ-type with EF hand domain 1 |
| chr23_+_25201077 | 0.11 |
ENSDART00000136675
|
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr2_+_111919 | 0.11 |
ENSDART00000149391
|
fggy
|
FGGY carbohydrate kinase domain containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of snai2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.2 | 0.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 0.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 0.6 | GO:0060283 | negative regulation of oocyte development(GO:0060283) |
| 0.1 | 1.3 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 0.5 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 1.3 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.4 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.1 | 0.6 | GO:0090133 | mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.1 | 0.3 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 0.3 | GO:0046689 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.1 | 0.8 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.1 | 0.5 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.4 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.3 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 0.3 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 0.9 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.1 | 0.5 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.5 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.5 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.1 | 0.2 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.5 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.4 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.7 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.5 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.4 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.4 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.7 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0033605 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.0 | 0.1 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.4 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.1 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.7 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 1.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.1 | 0.7 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.4 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 1.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.4 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.5 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.6 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.9 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.5 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.5 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.4 | GO:0043296 | apical junction complex(GO:0043296) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 1.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.6 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 0.9 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 1.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.2 | GO:0034618 | arginine binding(GO:0034618) |
| 0.1 | 0.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.3 | GO:0010858 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.6 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.7 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.5 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.5 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.0 | 0.2 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.7 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.7 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.3 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.4 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |