Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
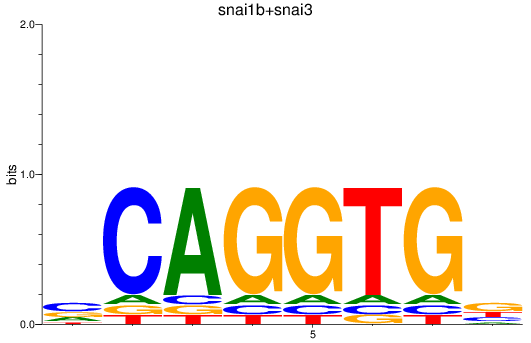
Results for snai1b+snai3
Z-value: 0.58
Transcription factors associated with snai1b+snai3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai3
|
ENSDARG00000031243 | snail family zinc finger 3 |
|
snai1b
|
ENSDARG00000046019 | snail family zinc finger 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai1b | dr11_v1_chr23_-_2513300_2513300 | -0.88 | 1.7e-06 | Click! |
| snai3 | dr11_v1_chr7_+_55112922_55112922 | 0.47 | 4.9e-02 | Click! |
Activity profile of snai1b+snai3 motif
Sorted Z-values of snai1b+snai3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_1052591 | 2.38 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr7_+_58751504 | 2.28 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr21_-_43666420 | 1.60 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr13_-_21672131 | 1.56 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr6_-_37749711 | 1.50 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr14_+_15155684 | 1.23 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr20_-_43743700 | 1.20 |
ENSDART00000100620
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr5_+_6670945 | 1.18 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr18_+_45666489 | 1.12 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr17_+_1360192 | 1.08 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr5_-_23696926 | 1.05 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr21_-_35325466 | 1.01 |
ENSDART00000134780
ENSDART00000145930 ENSDART00000076715 ENSDART00000065341 ENSDART00000162189 |
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr22_-_17677947 | 1.01 |
ENSDART00000139911
|
tjp3
|
tight junction protein 3 |
| chr2_+_26237322 | 0.96 |
ENSDART00000030520
|
palm1b
|
paralemmin 1b |
| chr13_-_36525982 | 0.91 |
ENSDART00000114744
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr10_+_37173029 | 0.91 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr12_+_16953415 | 0.84 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr14_-_31087830 | 0.81 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr9_-_28255029 | 0.77 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr20_-_23426339 | 0.75 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr13_+_2448251 | 0.74 |
ENSDART00000188361
|
arfgef3
|
ARFGEF family member 3 |
| chr16_+_41060161 | 0.69 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr11_+_6902946 | 0.69 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr15_+_784149 | 0.67 |
ENSDART00000155114
|
znf970
|
zinc finger protein 970 |
| chr5_-_22052852 | 0.67 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr18_+_6641542 | 0.66 |
ENSDART00000160379
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr5_-_12031174 | 0.63 |
ENSDART00000159896
|
castor1
|
cytosolic arginine sensor for mTORC1 subunit 1 |
| chr24_+_36018164 | 0.61 |
ENSDART00000182815
ENSDART00000126941 |
gnal2
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type 2 |
| chr11_-_43473824 | 0.59 |
ENSDART00000179561
|
tmem63bb
|
transmembrane protein 63Bb |
| chr6_-_10728921 | 0.58 |
ENSDART00000151484
|
sp3b
|
Sp3b transcription factor |
| chr13_+_2894536 | 0.57 |
ENSDART00000183678
|
CABZ01087629.1
|
|
| chr7_-_30926030 | 0.49 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr13_-_39254 | 0.49 |
ENSDART00000093222
|
gtf2a1l
|
general transcription factor IIA, 1-like |
| chr17_+_24590177 | 0.44 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr6_-_10728057 | 0.43 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr1_+_10019653 | 0.42 |
ENSDART00000190227
|
trim2b
|
tripartite motif containing 2b |
| chr7_+_20917966 | 0.42 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr10_-_38468847 | 0.41 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr14_-_15155384 | 0.41 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr22_+_22438783 | 0.40 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr20_-_52902693 | 0.37 |
ENSDART00000166115
ENSDART00000161050 |
ctsbb
|
cathepsin Bb |
| chr14_-_26482096 | 0.37 |
ENSDART00000187280
|
smad5
|
SMAD family member 5 |
| chr21_-_21465111 | 0.35 |
ENSDART00000141487
|
nectin3b
|
nectin cell adhesion molecule 3b |
| chr7_+_30988570 | 0.33 |
ENSDART00000180613
ENSDART00000185625 |
tjp1a
|
tight junction protein 1a |
| chr3_-_42016693 | 0.31 |
ENSDART00000184741
|
ttyh3a
|
tweety family member 3a |
| chr13_-_11800281 | 0.31 |
ENSDART00000079392
|
zmp:0000000662
|
zmp:0000000662 |
| chr22_-_18546649 | 0.30 |
ENSDART00000171277
|
cirbpb
|
cold inducible RNA binding protein b |
| chr8_-_10949847 | 0.30 |
ENSDART00000123209
|
pqlc2
|
PQ loop repeat containing 2 |
| chr1_-_55058795 | 0.26 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr20_-_21672970 | 0.26 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr17_+_50655619 | 0.25 |
ENSDART00000184917
ENSDART00000167638 |
ddhd1a
|
DDHD domain containing 1a |
| chr24_+_19578935 | 0.25 |
ENSDART00000137175
|
sulf1
|
sulfatase 1 |
| chr13_+_35955562 | 0.25 |
ENSDART00000137377
|
si:ch211-67f13.7
|
si:ch211-67f13.7 |
| chr19_+_142270 | 0.24 |
ENSDART00000160201
|
tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b |
| chr15_-_434503 | 0.23 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr17_+_24936059 | 0.23 |
ENSDART00000082438
|
dlgap2a
|
discs, large (Drosophila) homolog-associated protein 2a |
| chr7_+_38962459 | 0.23 |
ENSDART00000173851
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr8_+_20140321 | 0.23 |
ENSDART00000012120
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr13_-_9119867 | 0.21 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr2_+_5948534 | 0.18 |
ENSDART00000124324
ENSDART00000176461 |
slc1a7a
|
solute carrier family 1 (glutamate transporter), member 7a |
| chr17_-_11151655 | 0.17 |
ENSDART00000156383
|
CU179699.1
|
|
| chr8_+_247163 | 0.17 |
ENSDART00000122378
|
cep120
|
centrosomal protein 120 |
| chr8_+_8196087 | 0.17 |
ENSDART00000026965
|
plxnb3
|
plexin B3 |
| chr20_-_9436521 | 0.16 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr12_-_2869565 | 0.15 |
ENSDART00000152772
|
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr3_+_14641962 | 0.15 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr22_-_3595439 | 0.14 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr5_-_65159258 | 0.14 |
ENSDART00000160429
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr12_+_48784731 | 0.13 |
ENSDART00000158348
|
zmiz1b
|
zinc finger, MIZ-type containing 1b |
| chr3_+_23029934 | 0.12 |
ENSDART00000110343
|
nags
|
N-acetylglutamate synthase |
| chr1_-_43892349 | 0.12 |
ENSDART00000148416
|
tacr3a
|
tachykinin receptor 3a |
| chr7_+_42328578 | 0.10 |
ENSDART00000149082
|
phkb
|
phosphorylase kinase, beta |
| chr10_-_94184 | 0.08 |
ENSDART00000146125
|
ttc3
|
tetratricopeptide repeat domain 3 |
| chr2_-_42628028 | 0.08 |
ENSDART00000179866
|
myo10
|
myosin X |
| chr4_+_54645654 | 0.06 |
ENSDART00000192864
|
si:ch211-227e10.1
|
si:ch211-227e10.1 |
| chr4_-_36476889 | 0.05 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr18_-_5781922 | 0.03 |
ENSDART00000128722
|
RGS9BP
|
si:ch73-167i17.6 |
| chr2_-_722156 | 0.03 |
ENSDART00000045770
ENSDART00000169498 |
foxq1a
|
forkhead box Q1a |
| chr14_+_34514336 | 0.02 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr7_-_58867188 | 0.02 |
ENSDART00000187006
|
CU681855.1
|
|
| chr12_-_46112892 | 0.01 |
ENSDART00000187128
ENSDART00000114268 |
zgc:153932
|
zgc:153932 |
| chr10_+_24445698 | 0.01 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr4_-_16341801 | 0.00 |
ENSDART00000140190
|
kera
|
keratocan |
| chr1_+_524717 | 0.00 |
ENSDART00000102421
ENSDART00000184473 |
mrpl16
|
mitochondrial ribosomal protein L16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of snai1b+snai3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.2 | 0.8 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.1 | 0.7 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 1.0 | GO:0030104 | water homeostasis(GO:0030104) |
| 0.1 | 0.7 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.4 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.4 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.6 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.5 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.1 | 0.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.7 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.4 | GO:0035188 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.6 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.7 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.4 | GO:2000223 | regulation of BMP signaling pathway involved in heart jogging(GO:2000223) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:1902093 | positive regulation of sperm motility(GO:1902093) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.2 | GO:0098962 | regulation of postsynaptic neurotransmitter receptor activity(GO:0098962) |
| 0.0 | 0.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.4 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.1 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 2.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.7 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 1.7 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0034618 | arginine binding(GO:0034618) |
| 0.2 | 2.4 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 0.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.7 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.7 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 1.6 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.7 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 2.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.1 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.6 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.7 | GO:0032934 | sterol binding(GO:0032934) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 0.8 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |