Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
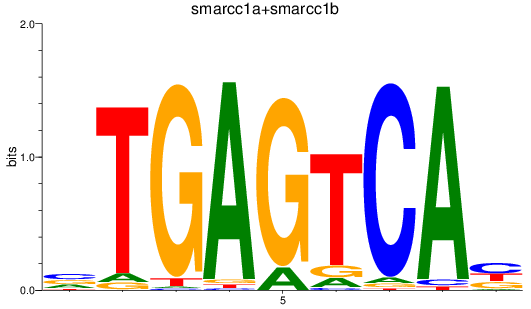
Results for smarcc1a+smarcc1b
Z-value: 0.42
Transcription factors associated with smarcc1a+smarcc1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smarcc1a
|
ENSDARG00000017397 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a |
|
smarcc1b
|
ENSDARG00000098919 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smarcc1a | dr11_v1_chr16_-_42461263_42461263 | 0.55 | 1.7e-02 | Click! |
| smarcc1b | dr11_v1_chr19_-_19379084_19379084 | 0.39 | 1.1e-01 | Click! |
Activity profile of smarcc1a+smarcc1b motif
Sorted Z-values of smarcc1a+smarcc1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_551963 | 0.55 |
ENSDART00000110495
|
akap9
|
A kinase (PRKA) anchor protein 9 |
| chr18_+_3243292 | 0.45 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr16_-_24832038 | 0.44 |
ENSDART00000153731
|
si:dkey-79d12.5
|
si:dkey-79d12.5 |
| chr16_+_23403602 | 0.43 |
ENSDART00000159848
|
s100w
|
S100 calcium binding protein W |
| chr22_+_11756040 | 0.41 |
ENSDART00000105808
|
krt97
|
keratin 97 |
| chr21_+_20901505 | 0.40 |
ENSDART00000132741
|
c7b
|
complement component 7b |
| chr2_-_985417 | 0.40 |
ENSDART00000140540
|
si:ch211-241e1.3
|
si:ch211-241e1.3 |
| chr20_+_28364742 | 0.38 |
ENSDART00000103355
|
rhov
|
ras homolog family member V |
| chr1_-_59252973 | 0.34 |
ENSDART00000167061
|
si:ch1073-286c18.5
|
si:ch1073-286c18.5 |
| chr1_+_46598764 | 0.31 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr25_+_18583877 | 0.31 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr21_+_20903244 | 0.30 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr5_-_49951106 | 0.29 |
ENSDART00000135954
|
fam172a
|
family with sequence similarity 172, member A |
| chr10_-_32877348 | 0.27 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr16_+_13818500 | 0.27 |
ENSDART00000135245
|
flcn
|
folliculin |
| chr5_-_42180205 | 0.25 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr22_+_8979955 | 0.25 |
ENSDART00000144005
|
si:ch211-213a13.1
|
si:ch211-213a13.1 |
| chr23_-_270847 | 0.24 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr14_+_24845941 | 0.24 |
ENSDART00000187513
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr19_-_5332784 | 0.24 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr12_-_3077395 | 0.23 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_+_13818743 | 0.23 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr3_-_40232615 | 0.23 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr2_-_898899 | 0.22 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr4_+_5317483 | 0.22 |
ENSDART00000150366
|
si:ch211-214j24.10
|
si:ch211-214j24.10 |
| chr2_+_36015049 | 0.22 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr21_+_25765734 | 0.22 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr6_-_8244474 | 0.22 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr2_-_39759059 | 0.22 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr25_+_7423770 | 0.21 |
ENSDART00000155458
|
ubap1la
|
ubiquitin associated protein 1-like a |
| chr8_+_19548985 | 0.21 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr17_-_35076730 | 0.21 |
ENSDART00000146590
|
mboat2a
|
membrane bound O-acyltransferase domain containing 2a |
| chr3_+_35498119 | 0.21 |
ENSDART00000178963
|
tnrc6a
|
trinucleotide repeat containing 6a |
| chr15_-_2657508 | 0.20 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr22_+_17399124 | 0.20 |
ENSDART00000145769
|
rabgap1l
|
RAB GTPase activating protein 1-like |
| chr2_+_25378457 | 0.20 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr19_+_48359259 | 0.20 |
ENSDART00000167353
|
sgo1
|
shugoshin 1 |
| chr5_-_50992690 | 0.20 |
ENSDART00000149553
ENSDART00000097460 ENSDART00000192021 |
hmgcra
|
3-hydroxy-3-methylglutaryl-CoA reductase a |
| chr21_+_10866421 | 0.19 |
ENSDART00000137858
|
alpk2
|
alpha-kinase 2 |
| chr22_+_11775269 | 0.19 |
ENSDART00000140272
|
krt96
|
keratin 96 |
| chr25_-_22187397 | 0.19 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr22_+_797105 | 0.19 |
ENSDART00000128549
|
cry1bb
|
cryptochrome circadian clock 1bb |
| chr7_-_60351537 | 0.18 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr17_+_51744450 | 0.18 |
ENSDART00000190955
ENSDART00000149807 |
odc1
|
ornithine decarboxylase 1 |
| chr12_+_23991639 | 0.18 |
ENSDART00000003143
|
psme4b
|
proteasome activator subunit 4b |
| chr5_+_24089334 | 0.18 |
ENSDART00000183748
|
tp53
|
tumor protein p53 |
| chr25_+_20119466 | 0.18 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr5_-_67629263 | 0.18 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr9_+_33154841 | 0.18 |
ENSDART00000132465
|
dopey2
|
dopey family member 2 |
| chr12_-_17147473 | 0.18 |
ENSDART00000106035
|
stambpl1
|
STAM binding protein-like 1 |
| chr2_+_48288461 | 0.18 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr12_-_27031060 | 0.18 |
ENSDART00000076103
|
chmp2a
|
charged multivesicular body protein 2A |
| chr14_+_24840669 | 0.17 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr14_+_22397251 | 0.17 |
ENSDART00000185239
ENSDART00000124072 ENSDART00000054977 |
atp7a
|
ATPase copper transporting alpha |
| chr7_+_26649319 | 0.17 |
ENSDART00000173823
ENSDART00000101053 |
tp53i11a
|
tumor protein p53 inducible protein 11a |
| chr6_-_60079551 | 0.17 |
ENSDART00000154753
|
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_+_66884570 | 0.17 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr1_+_46598502 | 0.17 |
ENSDART00000132861
|
cab39l
|
calcium binding protein 39-like |
| chr9_+_41024973 | 0.17 |
ENSDART00000014660
ENSDART00000144467 |
ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr7_-_26262978 | 0.17 |
ENSDART00000137769
|
ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr10_+_16225117 | 0.17 |
ENSDART00000169885
|
slc12a2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr25_+_14697247 | 0.17 |
ENSDART00000180747
|
mpped2
|
metallophosphoesterase domain containing 2b |
| chr24_-_33366188 | 0.16 |
ENSDART00000074161
|
slc4a2b
|
solute carrier family 4 (anion exchanger), member 2b |
| chr5_-_25582721 | 0.16 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr20_+_38837238 | 0.16 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr9_+_38481780 | 0.16 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr19_+_37120491 | 0.16 |
ENSDART00000032341
|
pef1
|
penta-EF-hand domain containing 1 |
| chr5_+_63767376 | 0.16 |
ENSDART00000138898
|
rgs3b
|
regulator of G protein signaling 3b |
| chr18_-_12957451 | 0.16 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr4_+_6840223 | 0.16 |
ENSDART00000161329
|
dock4b
|
dedicator of cytokinesis 4b |
| chr18_-_37355666 | 0.16 |
ENSDART00000098914
|
yap1
|
Yes-associated protein 1 |
| chr2_+_3516913 | 0.16 |
ENSDART00000109346
|
CU693445.1
|
|
| chr18_+_34599315 | 0.16 |
ENSDART00000159306
ENSDART00000088675 |
tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr10_-_2971407 | 0.16 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr7_-_9674073 | 0.15 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr3_+_50201240 | 0.15 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr9_+_28140089 | 0.15 |
ENSDART00000046880
|
plekhm3
|
pleckstrin homology domain containing, family M, member 3 |
| chr8_+_8671229 | 0.15 |
ENSDART00000131963
|
usp11
|
ubiquitin specific peptidase 11 |
| chr5_+_65086856 | 0.15 |
ENSDART00000169209
ENSDART00000162409 |
ptrh1
|
peptidyl-tRNA hydrolase 1 homolog |
| chr19_+_26072624 | 0.15 |
ENSDART00000147627
|
jarid2b
|
jumonji, AT rich interactive domain 2b |
| chr18_+_15841449 | 0.15 |
ENSDART00000141800
ENSDART00000091349 |
eea1
|
early endosome antigen 1 |
| chr7_-_20241346 | 0.15 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr7_+_34506937 | 0.15 |
ENSDART00000111303
|
rfx7a
|
regulatory factor X7a |
| chr19_-_7043355 | 0.14 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr4_+_25181572 | 0.14 |
ENSDART00000078529
ENSDART00000136643 |
kin
|
Kin17 DNA and RNA binding protein |
| chr14_+_23184517 | 0.14 |
ENSDART00000181410
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr8_+_53311965 | 0.14 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr18_+_3169579 | 0.14 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr10_+_32050906 | 0.14 |
ENSDART00000137373
|
si:ch211-266i6.3
|
si:ch211-266i6.3 |
| chr16_+_11242443 | 0.14 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr7_-_44963154 | 0.14 |
ENSDART00000073735
|
rrad
|
Ras-related associated with diabetes |
| chr2_-_27619954 | 0.14 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr7_-_60351876 | 0.14 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr15_+_15479442 | 0.14 |
ENSDART00000067095
|
nek8
|
NIMA-related kinase 8 |
| chr11_-_45420212 | 0.14 |
ENSDART00000182042
ENSDART00000163185 |
ankrd13c
|
ankyrin repeat domain 13C |
| chr8_-_11170114 | 0.14 |
ENSDART00000133532
|
si:ch211-204d2.4
|
si:ch211-204d2.4 |
| chr25_-_22191733 | 0.13 |
ENSDART00000067478
|
pkp3a
|
plakophilin 3a |
| chr5_-_23464970 | 0.13 |
ENSDART00000141028
|
si:dkeyp-20g2.1
|
si:dkeyp-20g2.1 |
| chr14_+_15768942 | 0.13 |
ENSDART00000158998
|
ergic1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr21_+_38745094 | 0.13 |
ENSDART00000113316
|
heatr6
|
HEAT repeat containing 6 |
| chr6_+_44197099 | 0.13 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr2_-_26642831 | 0.13 |
ENSDART00000132854
ENSDART00000087714 ENSDART00000132651 |
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr15_-_1844048 | 0.13 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr10_-_74408 | 0.13 |
ENSDART00000100073
ENSDART00000141723 |
dyrk1aa
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, a |
| chr16_+_42829735 | 0.13 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr7_-_5487593 | 0.13 |
ENSDART00000136594
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr22_+_23359369 | 0.13 |
ENSDART00000170886
|
dennd1b
|
DENN/MADD domain containing 1B |
| chr17_+_58211 | 0.13 |
ENSDART00000157642
|
si:ch1073-209e23.1
|
si:ch1073-209e23.1 |
| chr15_+_44201056 | 0.13 |
ENSDART00000162433
ENSDART00000148336 |
CU655961.4
|
|
| chr6_+_44197348 | 0.13 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr7_+_35068036 | 0.13 |
ENSDART00000022139
|
zgc:136461
|
zgc:136461 |
| chr20_+_23501535 | 0.12 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr12_-_7639120 | 0.12 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr13_-_37474989 | 0.12 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr21_+_45717930 | 0.12 |
ENSDART00000164315
|
ddx46
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 |
| chr5_-_23485161 | 0.12 |
ENSDART00000170293
ENSDART00000134069 |
si:dkeyp-20g2.1
si:dkeyp-20g2.3
|
si:dkeyp-20g2.1 si:dkeyp-20g2.3 |
| chr21_+_37513488 | 0.12 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr4_-_1757460 | 0.12 |
ENSDART00000144074
|
tm7sf3
|
transmembrane 7 superfamily member 3 |
| chr16_+_32136550 | 0.12 |
ENSDART00000147526
|
sphk2
|
sphingosine kinase 2 |
| chr15_-_19772372 | 0.12 |
ENSDART00000152729
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr16_+_54210554 | 0.12 |
ENSDART00000172622
|
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr16_-_2390931 | 0.12 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr6_+_59642695 | 0.12 |
ENSDART00000166373
ENSDART00000161030 |
R3HDM2
|
R3H domain containing 2 |
| chr15_-_31177324 | 0.12 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr15_+_24704798 | 0.12 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr20_+_34770197 | 0.12 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr17_+_15674052 | 0.12 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr20_-_14462995 | 0.12 |
ENSDART00000152418
ENSDART00000044125 |
grcc10
|
gene rich cluster, C10 gene |
| chr21_+_6328801 | 0.12 |
ENSDART00000163577
|
fnbp1b
|
formin binding protein 1b |
| chr19_+_15440841 | 0.12 |
ENSDART00000182329
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr11_-_29936755 | 0.12 |
ENSDART00000145251
|
pir
|
pirin |
| chr25_-_3808635 | 0.11 |
ENSDART00000075659
ENSDART00000154691 |
gatd1
|
glutamine amidotransferase like class 1 domain containing 1 |
| chr6_+_11397269 | 0.11 |
ENSDART00000114260
|
senp2
|
SUMO1/sentrin/SMT3 specific peptidase 2 |
| chr15_-_19771981 | 0.11 |
ENSDART00000175502
ENSDART00000159475 |
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr19_+_23932259 | 0.11 |
ENSDART00000139040
|
si:dkey-222b8.1
|
si:dkey-222b8.1 |
| chr12_+_32292564 | 0.11 |
ENSDART00000152945
|
ANKFN1
|
si:ch211-277e21.2 |
| chr10_+_22771176 | 0.11 |
ENSDART00000192046
|
tmem88a
|
transmembrane protein 88 a |
| chr21_+_13327527 | 0.11 |
ENSDART00000114294
|
snrpd3l
|
small nuclear ribonucleoprotein D3 polypeptide, like |
| chr23_-_14937101 | 0.11 |
ENSDART00000010258
|
ndrg3b
|
ndrg family member 3b |
| chr1_+_51039558 | 0.11 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr1_-_34750169 | 0.11 |
ENSDART00000149380
|
si:dkey-151m22.8
|
si:dkey-151m22.8 |
| chr8_-_16559181 | 0.11 |
ENSDART00000098692
ENSDART00000124257 |
eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr7_-_7764287 | 0.11 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr22_-_6562618 | 0.11 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr13_-_26799244 | 0.11 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr15_-_38129845 | 0.11 |
ENSDART00000057095
|
si:dkey-24p1.1
|
si:dkey-24p1.1 |
| chr10_+_5744941 | 0.11 |
ENSDART00000159769
ENSDART00000184734 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr13_+_7575563 | 0.11 |
ENSDART00000146592
|
gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr23_+_9268083 | 0.11 |
ENSDART00000055054
|
acss2
|
acyl-CoA synthetase short chain family member 2 |
| chr19_+_1510971 | 0.11 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr23_-_7799184 | 0.11 |
ENSDART00000190946
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr10_+_5234327 | 0.10 |
ENSDART00000133927
ENSDART00000063120 |
sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr5_-_21888368 | 0.10 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr23_-_35694461 | 0.10 |
ENSDART00000185884
|
tuba1c
|
tubulin, alpha 1c |
| chr9_+_33261330 | 0.10 |
ENSDART00000135384
|
usp9
|
ubiquitin specific peptidase 9 |
| chr2_+_31437547 | 0.10 |
ENSDART00000141170
|
stam
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 |
| chr2_+_15069011 | 0.10 |
ENSDART00000145893
|
cnn3b
|
calponin 3, acidic b |
| chr24_-_21973365 | 0.10 |
ENSDART00000081204
ENSDART00000030592 |
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr21_-_9914745 | 0.10 |
ENSDART00000172124
|
arhgap24
|
Rho GTPase activating protein 24 |
| chr2_+_37838259 | 0.10 |
ENSDART00000136796
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr2_-_32366287 | 0.10 |
ENSDART00000144758
|
ubtfl
|
upstream binding transcription factor, like |
| chr12_-_27212880 | 0.10 |
ENSDART00000002835
|
psme3
|
proteasome activator subunit 3 |
| chr14_-_34605804 | 0.10 |
ENSDART00000144547
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr15_+_40188076 | 0.10 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr16_-_13818061 | 0.10 |
ENSDART00000132982
ENSDART00000144856 |
leng9
|
leukocyte receptor cluster (LRC) member 9 |
| chr22_+_465269 | 0.10 |
ENSDART00000145767
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr7_+_34549198 | 0.10 |
ENSDART00000173784
|
fhod1
|
formin homology 2 domain containing 1 |
| chr22_-_881725 | 0.10 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr5_+_6670945 | 0.10 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr3_+_15358459 | 0.10 |
ENSDART00000141808
|
sh2b1
|
SH2B adaptor protein 1 |
| chr5_+_61556172 | 0.10 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr22_+_16320076 | 0.10 |
ENSDART00000164161
|
osbpl1a
|
oxysterol binding protein-like 1A |
| chr20_+_29209767 | 0.10 |
ENSDART00000141252
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr9_+_426392 | 0.10 |
ENSDART00000172515
|
bzw1b
|
basic leucine zipper and W2 domains 1b |
| chr22_-_26595027 | 0.10 |
ENSDART00000184162
|
CABZ01072309.1
|
|
| chr11_-_24681292 | 0.09 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr13_-_293250 | 0.09 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr1_-_40227166 | 0.09 |
ENSDART00000146680
|
si:ch211-113e8.3
|
si:ch211-113e8.3 |
| chr14_-_34605607 | 0.09 |
ENSDART00000191608
|
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr11_-_10770053 | 0.09 |
ENSDART00000179213
|
slc4a10a
|
solute carrier family 4, sodium bicarbonate transporter, member 10a |
| chr5_+_4533244 | 0.09 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr6_+_56147812 | 0.09 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr16_-_51288178 | 0.09 |
ENSDART00000079864
|
zgc:173729
|
zgc:173729 |
| chr2_+_10709557 | 0.09 |
ENSDART00000183118
ENSDART00000109723 |
evi5a
|
ecotropic viral integration site 5a |
| chr3_-_5644028 | 0.09 |
ENSDART00000019957
|
ddx39ab
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Ab |
| chr7_+_26762717 | 0.09 |
ENSDART00000186464
|
tspan18a
|
tetraspanin 18a |
| chr16_-_21915223 | 0.09 |
ENSDART00000170634
|
setdb1b
|
SET domain, bifurcated 1b |
| chr2_-_42864472 | 0.09 |
ENSDART00000134139
|
adcy8
|
adenylate cyclase 8 (brain) |
| chr9_+_7030016 | 0.09 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr10_-_7988396 | 0.09 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr21_-_30254185 | 0.09 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr20_+_29209926 | 0.09 |
ENSDART00000152949
ENSDART00000153016 |
katnbl1
|
katanin p80 subunit B-like 1 |
| chr11_+_25693395 | 0.09 |
ENSDART00000110224
|
mon1bb
|
MON1 secretory trafficking family member Bb |
| chr16_-_13613475 | 0.09 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr2_-_49557932 | 0.09 |
ENSDART00000141902
|
si:ch211-209f23.3
|
si:ch211-209f23.3 |
| chr13_+_7241170 | 0.09 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr8_+_21229718 | 0.09 |
ENSDART00000100222
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr14_-_38889840 | 0.09 |
ENSDART00000035779
|
zgc:101583
|
zgc:101583 |
| chr24_-_24060460 | 0.09 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr13_-_33134611 | 0.09 |
ENSDART00000026280
|
plek2
|
pleckstrin 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of smarcc1a+smarcc1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.6 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.2 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.2 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.1 | 0.5 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.1 | 0.2 | GO:0010526 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.2 | GO:0071962 | mitotic sister chromatid cohesion, centromeric(GO:0071962) |
| 0.0 | 0.1 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.3 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.0 | 0.2 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.1 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 0.1 | GO:0055026 | negative regulation of striated muscle cell differentiation(GO:0051154) negative regulation of cardiac muscle tissue development(GO:0055026) cardiac muscle cell fate commitment(GO:0060923) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) negative regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901296) negative regulation of cardiocyte differentiation(GO:1905208) negative regulation of cardiac muscle cell differentiation(GO:2000726) |
| 0.0 | 0.1 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.2 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.0 | 0.2 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.1 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.3 | GO:0021707 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0045628 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.7 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.3 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.3 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.2 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.3 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.0 | 0.1 | GO:0006031 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 | 0.1 | GO:0048939 | anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.3 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:1900028 | wound healing, spreading of epidermal cells(GO:0035313) negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.0 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.0 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0015824 | proline transport(GO:0015824) |
| 0.0 | 0.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.3 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0017003 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0048385 | regulation of retinoic acid receptor signaling pathway(GO:0048385) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.2 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0098894 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.5 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.1 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.0 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.1 | 0.3 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0017050 | D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0009384 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.0 | 0.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.2 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0005183 | gonadotropin hormone-releasing hormone activity(GO:0005183) gonadotropin-releasing hormone receptor binding(GO:0031530) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.0 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.0 | 0.1 | GO:0004586 | ornithine decarboxylase activity(GO:0004586) |
| 0.0 | 0.0 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.1 | 0.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.1 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |