Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
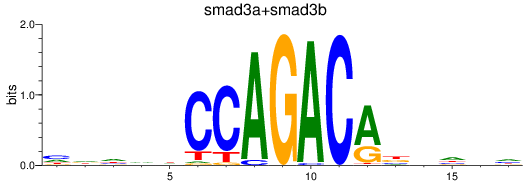
Results for smad3a+smad3b
Z-value: 0.63
Transcription factors associated with smad3a+smad3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smad3b
|
ENSDARG00000010207 | SMAD family member 3b |
|
smad3a
|
ENSDARG00000036096 | SMAD family member 3a |
|
smad3a
|
ENSDARG00000117146 | SMAD family member 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smad3a | dr11_v1_chr7_-_34149263_34149263 | -0.77 | 2.1e-04 | Click! |
| smad3b | dr11_v1_chr18_+_19772874_19772874 | -0.54 | 2.2e-02 | Click! |
Activity profile of smad3a+smad3b motif
Sorted Z-values of smad3a+smad3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_32204470 | 0.78 |
ENSDART00000143928
|
rwdd1
|
RWD domain containing 1 |
| chr19_+_627899 | 0.77 |
ENSDART00000148508
|
tert
|
telomerase reverse transcriptase |
| chr2_+_54641644 | 0.77 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr8_+_39760258 | 0.76 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr3_+_13603272 | 0.71 |
ENSDART00000185084
|
hspbp1
|
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr11_-_8208464 | 0.65 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr19_-_8962884 | 0.64 |
ENSDART00000172582
ENSDART00000104657 |
mrps21
|
mitochondrial ribosomal protein S21 |
| chr12_+_4712215 | 0.63 |
ENSDART00000152134
|
kansl1a
|
KAT8 regulatory NSL complex subunit 1a |
| chr20_-_4883673 | 0.63 |
ENSDART00000145540
ENSDART00000053877 |
zdhhc14
|
zinc finger, DHHC-type containing 14 |
| chr5_+_62723233 | 0.62 |
ENSDART00000183718
|
nanos2
|
nanos homolog 2 |
| chr2_-_20599315 | 0.62 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr21_+_26071874 | 0.60 |
ENSDART00000003001
ENSDART00000146573 |
rpl23a
|
ribosomal protein L23a |
| chr23_+_44614056 | 0.55 |
ENSDART00000188379
|
eno3
|
enolase 3, (beta, muscle) |
| chr13_-_3370638 | 0.55 |
ENSDART00000029649
|
prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr24_-_37698796 | 0.52 |
ENSDART00000172178
|
si:ch211-231f6.6
|
si:ch211-231f6.6 |
| chr9_-_21369765 | 0.51 |
ENSDART00000132032
|
ift88
|
intraflagellar transport 88 homolog |
| chr22_-_28871787 | 0.50 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr22_+_36914636 | 0.50 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr3_-_10779777 | 0.49 |
ENSDART00000153911
|
znf1004
|
zinc finger protein 1004 |
| chr5_+_71999996 | 0.48 |
ENSDART00000179933
ENSDART00000187070 |
PLPP7 (1 of many)
|
phospholipid phosphatase 7 (inactive) |
| chr4_-_11737939 | 0.44 |
ENSDART00000150299
|
podxl
|
podocalyxin-like |
| chr3_-_58644920 | 0.44 |
ENSDART00000155953
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr20_-_27325258 | 0.43 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr16_+_53603945 | 0.43 |
ENSDART00000083513
|
sacm1lb
|
SAC1 like phosphatidylinositide phosphatase b |
| chr23_-_35694461 | 0.43 |
ENSDART00000185884
|
tuba1c
|
tubulin, alpha 1c |
| chr21_-_26071773 | 0.42 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr7_-_1504382 | 0.41 |
ENSDART00000172770
|
si:zfos-405g10.4
|
si:zfos-405g10.4 |
| chr8_-_23194408 | 0.41 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr23_+_13928346 | 0.40 |
ENSDART00000155326
|
si:dkey-90a13.10
|
si:dkey-90a13.10 |
| chr16_+_19013257 | 0.39 |
ENSDART00000144102
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr5_-_31716713 | 0.38 |
ENSDART00000131443
|
dpm2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr21_+_25533531 | 0.37 |
ENSDART00000134052
|
nlrc3l1
|
NLR family, CARD domain containing 3-like 1 |
| chr1_+_52563298 | 0.37 |
ENSDART00000142465
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr21_+_25533908 | 0.37 |
ENSDART00000185225
|
nlrc3l1
|
NLR family, CARD domain containing 3-like 1 |
| chr1_-_57172294 | 0.37 |
ENSDART00000063774
|
rac1l
|
Rac family small GTPase 1, like |
| chr5_+_13385837 | 0.36 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr7_+_50448478 | 0.35 |
ENSDART00000098815
|
serinc4
|
serine incorporator 4 |
| chr20_-_43495506 | 0.35 |
ENSDART00000170679
|
pimr132
|
Pim proto-oncogene, serine/threonine kinase, related 132 |
| chr21_+_20903244 | 0.35 |
ENSDART00000186193
|
c7b
|
complement component 7b |
| chr13_+_17694845 | 0.35 |
ENSDART00000079778
|
ifit8
|
interferon-induced protein with tetratricopeptide repeats 8 |
| chr3_-_28872756 | 0.34 |
ENSDART00000138840
|
eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr20_-_29683754 | 0.34 |
ENSDART00000130599
ENSDART00000015928 ENSDART00000131219 |
si:ch211-195d17.2
|
si:ch211-195d17.2 |
| chr7_+_29053217 | 0.34 |
ENSDART00000134332
ENSDART00000137241 |
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr25_-_35126024 | 0.33 |
ENSDART00000185109
|
HIST1H4I
|
zgc:165555 |
| chr2_-_7605121 | 0.33 |
ENSDART00000182099
|
CABZ01021599.1
|
|
| chr7_-_73834812 | 0.33 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr14_-_28052474 | 0.33 |
ENSDART00000172948
ENSDART00000135337 |
TSC22D3 (1 of many)
zgc:64189
|
si:ch211-220e11.3 zgc:64189 |
| chr4_-_11577253 | 0.32 |
ENSDART00000144452
|
net1
|
neuroepithelial cell transforming 1 |
| chr14_+_38786298 | 0.31 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr4_+_16787488 | 0.31 |
ENSDART00000143006
|
golt1ba
|
golgi transport 1Ba |
| chr4_-_14192254 | 0.30 |
ENSDART00000143804
|
pus7l
|
pseudouridylate synthase 7-like |
| chr22_-_10591876 | 0.30 |
ENSDART00000105846
|
si:dkey-42i9.8
|
si:dkey-42i9.8 |
| chr9_-_7378566 | 0.29 |
ENSDART00000144003
|
slc23a3
|
solute carrier family 23, member 3 |
| chr4_-_73709502 | 0.29 |
ENSDART00000170308
|
BX855614.3
|
|
| chr12_-_17479078 | 0.29 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr15_+_2500510 | 0.29 |
ENSDART00000180329
|
wdr53
|
WD repeat domain 53 |
| chr9_-_16853462 | 0.28 |
ENSDART00000160273
|
CT573248.2
|
|
| chr12_-_20795867 | 0.28 |
ENSDART00000152835
ENSDART00000153424 |
nme2a
|
NME/NM23 nucleoside diphosphate kinase 2a |
| chr14_+_6546516 | 0.28 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr7_+_66822229 | 0.28 |
ENSDART00000112109
|
lyve1a
|
lymphatic vessel endothelial hyaluronic receptor 1a |
| chr1_+_54043563 | 0.27 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr25_+_35056619 | 0.27 |
ENSDART00000154389
|
HIST1H4J
|
histone cluster 1 H4 family member j |
| chr7_+_21180747 | 0.26 |
ENSDART00000185543
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr21_+_18353703 | 0.25 |
ENSDART00000181396
ENSDART00000166359 |
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr2_-_22286828 | 0.25 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr1_+_21731382 | 0.24 |
ENSDART00000054395
|
pax5
|
paired box 5 |
| chr23_-_31396344 | 0.23 |
ENSDART00000112619
|
lca5
|
Leber congenital amaurosis 5 |
| chr25_+_35065802 | 0.23 |
ENSDART00000182372
|
zgc:165555
|
zgc:165555 |
| chr3_-_61203203 | 0.23 |
ENSDART00000171787
|
pvalb1
|
parvalbumin 1 |
| chr17_+_12285285 | 0.22 |
ENSDART00000154336
|
pimr174
|
Pim proto-oncogene, serine/threonine kinase, related 174 |
| chr22_+_36582963 | 0.22 |
ENSDART00000132181
|
armc9
|
armadillo repeat containing 9 |
| chr2_-_19520324 | 0.22 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr16_-_2807527 | 0.22 |
ENSDART00000148927
|
tgfb2
|
transforming growth factor, beta 2 |
| chr8_-_3346692 | 0.22 |
ENSDART00000057874
|
FUT9 (1 of many)
|
zgc:103510 |
| chr6_+_9289802 | 0.20 |
ENSDART00000188650
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr2_+_42236118 | 0.20 |
ENSDART00000114162
ENSDART00000141199 |
ftr02
|
finTRIM family, member 2 |
| chr1_-_59139599 | 0.20 |
ENSDART00000152233
|
si:ch1073-110a20.3
|
si:ch1073-110a20.3 |
| chr2_+_19578446 | 0.20 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr15_-_23793641 | 0.20 |
ENSDART00000122891
|
tmem97
|
transmembrane protein 97 |
| chr14_+_17003757 | 0.19 |
ENSDART00000170208
|
limch1b
|
LIM and calponin homology domains 1b |
| chr14_-_40797117 | 0.19 |
ENSDART00000122369
|
elf1
|
E74-like ETS transcription factor 1 |
| chr8_-_18203274 | 0.19 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr17_+_9009098 | 0.19 |
ENSDART00000180856
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr12_-_22509069 | 0.18 |
ENSDART00000179755
ENSDART00000109707 |
neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr3_-_62417677 | 0.18 |
ENSDART00000101888
|
sstr2b
|
somatostatin receptor 2b |
| chr4_-_18954001 | 0.18 |
ENSDART00000144814
|
si:dkey-31f5.8
|
si:dkey-31f5.8 |
| chr3_+_24190207 | 0.18 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr17_-_45009782 | 0.18 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr6_+_49926115 | 0.17 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr6_+_58698475 | 0.17 |
ENSDART00000056138
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr16_+_19013018 | 0.16 |
ENSDART00000191526
|
si:ch211-254p10.2
|
si:ch211-254p10.2 |
| chr23_+_42304602 | 0.16 |
ENSDART00000166004
|
cyp2aa11
|
cytochrome P450, family 2, subfamily AA, polypeptide 11 |
| chr8_+_23194212 | 0.16 |
ENSDART00000028946
ENSDART00000186842 ENSDART00000137815 ENSDART00000136914 ENSDART00000144855 ENSDART00000182689 |
tpd52l2a
|
tumor protein D52-like 2a |
| chr8_+_29267093 | 0.16 |
ENSDART00000077647
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr12_-_33805366 | 0.16 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr19_-_15281996 | 0.16 |
ENSDART00000103784
|
edn2
|
endothelin 2 |
| chr17_+_12265095 | 0.16 |
ENSDART00000153812
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr25_-_35956344 | 0.16 |
ENSDART00000066987
|
mphosph6
|
M-phase phosphoprotein 6 |
| chr16_+_5774977 | 0.15 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr11_+_45448212 | 0.15 |
ENSDART00000173341
|
sos1
|
son of sevenless homolog 1 (Drosophila) |
| chr11_+_12175162 | 0.15 |
ENSDART00000125446
|
si:ch211-156l18.7
|
si:ch211-156l18.7 |
| chr2_+_32796873 | 0.15 |
ENSDART00000077511
|
ccr9a
|
chemokine (C-C motif) receptor 9a |
| chr9_+_29616854 | 0.15 |
ENSDART00000033902
ENSDART00000143493 |
phf11
|
PHD finger protein 11 |
| chr7_-_69121896 | 0.15 |
ENSDART00000130227
|
crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr1_-_52494122 | 0.14 |
ENSDART00000131407
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr20_-_52199296 | 0.14 |
ENSDART00000131806
ENSDART00000143668 |
hlx1
|
H2.0-like homeo box 1 (Drosophila) |
| chr11_+_30310170 | 0.14 |
ENSDART00000127797
|
ugt1b3
|
UDP glucuronosyltransferase 1 family, polypeptide B3 |
| chr15_-_41734639 | 0.14 |
ENSDART00000154230
ENSDART00000167443 |
ftr90
|
finTRIM family, member 90 |
| chr21_-_17603182 | 0.14 |
ENSDART00000020048
ENSDART00000177270 |
gsna
|
gelsolin a |
| chr17_-_24564674 | 0.14 |
ENSDART00000105435
ENSDART00000135086 |
abch1
|
ATP-binding cassette, sub-family H, member 1 |
| chr1_-_39943596 | 0.14 |
ENSDART00000149730
|
stox2a
|
storkhead box 2a |
| chr3_-_58189429 | 0.14 |
ENSDART00000156092
|
si:ch211-256e16.11
|
si:ch211-256e16.11 |
| chr24_+_9744012 | 0.13 |
ENSDART00000129656
|
tmem108
|
transmembrane protein 108 |
| chr25_-_173165 | 0.13 |
ENSDART00000193594
|
CABZ01114053.1
|
|
| chr13_+_33298338 | 0.13 |
ENSDART00000131892
ENSDART00000143895 |
iqcc
|
IQ motif containing C |
| chr20_-_54377933 | 0.12 |
ENSDART00000182664
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr18_-_7454422 | 0.12 |
ENSDART00000124709
|
pdp2
|
putative pyruvate dehydrogenase phosphatase isoenzyme 2 |
| chr23_+_36052944 | 0.12 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr8_-_13541514 | 0.12 |
ENSDART00000063834
|
zgc:86586
|
zgc:86586 |
| chr17_+_14886828 | 0.12 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr8_-_52594111 | 0.12 |
ENSDART00000167667
|
si:ch73-199g24.2
|
si:ch73-199g24.2 |
| chr22_-_4398069 | 0.12 |
ENSDART00000181893
|
kdm4b
|
lysine (K)-specific demethylase 4B |
| chr24_-_6038025 | 0.11 |
ENSDART00000077819
ENSDART00000139216 |
ftr61
|
finTRIM family, member 61 |
| chr19_-_46775562 | 0.11 |
ENSDART00000187144
|
abrab
|
actin binding Rho activating protein b |
| chr6_-_50404732 | 0.11 |
ENSDART00000055510
|
romo1
|
reactive oxygen species modulator 1 |
| chr17_+_27134806 | 0.11 |
ENSDART00000151901
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr18_+_20560442 | 0.11 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr6_-_8498908 | 0.11 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr7_-_51528661 | 0.11 |
ENSDART00000174263
|
nhsl2
|
NHS-like 2 |
| chr19_+_4068134 | 0.11 |
ENSDART00000158285
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr13_+_10023256 | 0.11 |
ENSDART00000110035
|
srbd1
|
S1 RNA binding domain 1 |
| chr9_-_43375205 | 0.11 |
ENSDART00000138436
|
znf385b
|
zinc finger protein 385B |
| chr23_-_2099350 | 0.11 |
ENSDART00000091532
|
ndnf
|
neuron-derived neurotrophic factor |
| chr23_-_21453614 | 0.11 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr19_+_1097393 | 0.11 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr25_+_36315127 | 0.11 |
ENSDART00000191984
|
zgc:165555
|
zgc:165555 |
| chr1_+_24366110 | 0.11 |
ENSDART00000139913
|
pimr178
|
Pim proto-oncogene, serine/threonine kinase, related 178 |
| chr21_-_16399778 | 0.10 |
ENSDART00000136435
|
unc5da
|
unc-5 netrin receptor Da |
| chr25_-_29415369 | 0.10 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr24_+_36373588 | 0.10 |
ENSDART00000079101
|
map3k14b
|
mitogen-activated protein kinase kinase kinase 14b |
| chr9_-_41818760 | 0.10 |
ENSDART00000140601
|
si:dkeyp-30e7.2
|
si:dkeyp-30e7.2 |
| chr20_+_35282682 | 0.10 |
ENSDART00000187199
|
fam49a
|
family with sequence similarity 49, member A |
| chr13_-_31397987 | 0.10 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr12_+_4573696 | 0.10 |
ENSDART00000152534
|
si:dkey-94f20.4
|
si:dkey-94f20.4 |
| chr5_+_38685089 | 0.10 |
ENSDART00000139743
|
si:dkey-58f10.10
|
si:dkey-58f10.10 |
| chr25_-_20268027 | 0.10 |
ENSDART00000138763
|
dnajb9a
|
DnaJ (Hsp40) homolog, subfamily B, member 9a |
| chr11_+_8170948 | 0.10 |
ENSDART00000112127
ENSDART00000186267 |
dnase2b
|
deoxyribonuclease II beta |
| chr3_-_27784457 | 0.09 |
ENSDART00000019098
|
dnase1
|
deoxyribonuclease I |
| chr19_-_8604429 | 0.09 |
ENSDART00000151165
|
trim46b
|
tripartite motif containing 46b |
| chr7_+_2228276 | 0.09 |
ENSDART00000064294
|
si:dkey-187j14.4
|
si:dkey-187j14.4 |
| chr11_+_30306606 | 0.09 |
ENSDART00000128276
ENSDART00000190222 |
ugt1b4
|
UDP glucuronosyltransferase 1 family, polypeptide B4 |
| chr23_-_21463788 | 0.09 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr1_-_22861348 | 0.09 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr10_+_23441488 | 0.09 |
ENSDART00000100783
|
si:ch73-122g19.1
|
si:ch73-122g19.1 |
| chr2_+_47927026 | 0.09 |
ENSDART00000143023
|
ftr25
|
finTRIM family, member 25 |
| chr5_+_24287927 | 0.09 |
ENSDART00000143563
|
zdhhc23a
|
zinc finger, DHHC-type containing 23a |
| chr17_-_51679784 | 0.09 |
ENSDART00000103374
|
atp6v1c2
|
ATPase H+ transporting V1 subunit C2 |
| chr1_+_23784905 | 0.09 |
ENSDART00000171951
ENSDART00000188521 ENSDART00000183029 ENSDART00000187183 |
slit2
|
slit homolog 2 (Drosophila) |
| chr19_-_3167729 | 0.09 |
ENSDART00000110763
ENSDART00000145710 ENSDART00000074620 ENSDART00000105174 |
stm
|
starmaker |
| chr11_-_27874116 | 0.09 |
ENSDART00000180579
|
eif4g3a
|
eukaryotic translation initiation factor 4 gamma, 3a |
| chr1_+_59073203 | 0.08 |
ENSDART00000149937
ENSDART00000162201 |
MFAP4 (1 of many)
zgc:173915
|
si:zfos-2330d3.3 zgc:173915 |
| chr13_-_23264724 | 0.08 |
ENSDART00000051886
|
si:dkey-103j14.5
|
si:dkey-103j14.5 |
| chr4_+_33654247 | 0.08 |
ENSDART00000192537
|
si:dkey-84h14.1
|
si:dkey-84h14.1 |
| chr6_+_6491013 | 0.08 |
ENSDART00000140827
|
bcl11ab
|
B cell CLL/lymphoma 11Ab |
| chr12_-_44016898 | 0.08 |
ENSDART00000175304
|
si:dkey-201i2.4
|
si:dkey-201i2.4 |
| chr25_-_2081371 | 0.08 |
ENSDART00000104915
ENSDART00000156925 |
wnt7bb
|
wingless-type MMTV integration site family, member 7Bb |
| chr14_+_8504410 | 0.08 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| chr11_+_15890984 | 0.08 |
ENSDART00000158433
|
pank4
|
pantothenate kinase 4 |
| chr3_-_33029426 | 0.08 |
ENSDART00000181287
|
CT583642.1
|
|
| chr9_-_41153896 | 0.08 |
ENSDART00000059667
|
wdr75
|
WD repeat domain 75 |
| chr14_-_22385779 | 0.08 |
ENSDART00000192211
ENSDART00000175309 |
fbxl3l
|
F-box and leucine-rich repeat protein 3, like |
| chr25_+_36345867 | 0.08 |
ENSDART00000186527
|
zgc:165555
|
zgc:165555 |
| chr20_-_2619316 | 0.08 |
ENSDART00000185777
|
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr22_-_28373698 | 0.08 |
ENSDART00000157592
|
si:ch211-213c4.5
|
si:ch211-213c4.5 |
| chr20_+_46213553 | 0.08 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr18_+_13164325 | 0.08 |
ENSDART00000189057
|
tat
|
tyrosine aminotransferase |
| chr14_-_10762629 | 0.08 |
ENSDART00000061928
|
fgf16
|
fibroblast growth factor 16 |
| chr6_+_28051978 | 0.07 |
ENSDART00000143218
|
si:ch73-194h10.2
|
si:ch73-194h10.2 |
| chr3_+_52753416 | 0.07 |
ENSDART00000171185
|
gmip
|
GEM interacting protein |
| chr23_-_19230627 | 0.07 |
ENSDART00000007122
|
guca1b
|
guanylate cyclase activator 1B |
| chr20_+_46897504 | 0.07 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr10_-_24689725 | 0.07 |
ENSDART00000079566
|
si:ch211-287a12.9
|
si:ch211-287a12.9 |
| chr15_+_38007237 | 0.07 |
ENSDART00000182271
|
CR944667.2
|
|
| chr14_-_48348973 | 0.07 |
ENSDART00000185822
|
CABZ01080056.1
|
|
| chr6_-_8498676 | 0.07 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr1_-_30366824 | 0.07 |
ENSDART00000125759
|
si:dkey-22o12.2
|
si:dkey-22o12.2 |
| chr22_-_17499513 | 0.07 |
ENSDART00000105460
|
si:ch211-197g15.6
|
si:ch211-197g15.6 |
| chr10_-_24416471 | 0.07 |
ENSDART00000135985
|
hephl1b
|
hephaestin-like 1b |
| chr8_-_3336273 | 0.07 |
ENSDART00000081160
|
zgc:173737
|
zgc:173737 |
| chr19_-_10360188 | 0.07 |
ENSDART00000150933
|
si:ch211-232m10.6
|
si:ch211-232m10.6 |
| chr21_-_27362938 | 0.07 |
ENSDART00000131297
|
rin1a
|
Ras and Rab interactor 1a |
| chr24_-_33291784 | 0.07 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr2_-_59157790 | 0.07 |
ENSDART00000192303
ENSDART00000159362 |
ftr32
|
finTRIM family, member 32 |
| chr23_-_10914275 | 0.07 |
ENSDART00000112965
|
pdzrn3a
|
PDZ domain containing RING finger 3a |
| chr18_+_17611627 | 0.07 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr18_-_31227615 | 0.06 |
ENSDART00000003210
|
mc1r
|
melanocortin 1 receptor |
| chr19_+_7043634 | 0.06 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr13_+_2830574 | 0.06 |
ENSDART00000138143
|
si:ch211-233m11.1
|
si:ch211-233m11.1 |
| chr16_-_53259409 | 0.06 |
ENSDART00000157080
|
si:ch211-269k10.4
|
si:ch211-269k10.4 |
| chr10_-_36214582 | 0.06 |
ENSDART00000166471
|
or109-11
|
odorant receptor, family D, subfamily 109, member 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of smad3a+smad3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.2 | 0.5 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.1 | 0.4 | GO:0033632 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.1 | 0.3 | GO:0000103 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.4 | GO:0010874 | regulation of cholesterol efflux(GO:0010874) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 1.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.6 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.1 | 0.6 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.1 | 0.2 | GO:0016045 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0060584 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.3 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.1 | GO:1904184 | regulation of pyruvate dehydrogenase activity(GO:1904182) positive regulation of pyruvate dehydrogenase activity(GO:1904184) |
| 0.0 | 0.1 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0021543 | pallium development(GO:0021543) cerebral cortex cell migration(GO:0021795) cerebral cortex tangential migration(GO:0021800) cerebral cortex tangential migration using cell-axon interactions(GO:0021824) substrate-dependent cerebral cortex tangential migration(GO:0021825) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) cerebral cortex development(GO:0021987) |
| 0.0 | 0.2 | GO:0060118 | reflex(GO:0060004) vestibular reflex(GO:0060005) vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 0.0 | 0.6 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.0 | 0.2 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:2000379 | protein import into mitochondrial inner membrane(GO:0045039) positive regulation of reactive oxygen species metabolic process(GO:2000379) |
| 0.0 | 0.2 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.7 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.2 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.3 | GO:1900107 | regulation of nodal signaling pathway(GO:1900107) |
| 0.0 | 0.3 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.0 | 0.2 | GO:0051893 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 | 0.2 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.0 | GO:0046048 | ribonucleoside diphosphate catabolic process(GO:0009191) pyrimidine ribonucleoside diphosphate metabolic process(GO:0009193) UDP metabolic process(GO:0046048) |
| 0.0 | 0.0 | GO:0051228 | mitotic spindle disassembly(GO:0051228) spindle disassembly(GO:0051230) |
| 0.0 | 0.1 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.0 | 0.3 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.0 | 0.3 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.4 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.8 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.8 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.0 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 0.4 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.3 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.4 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.6 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.1 | 0.2 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.1 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0032038 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.2 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.8 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.0 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.8 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |