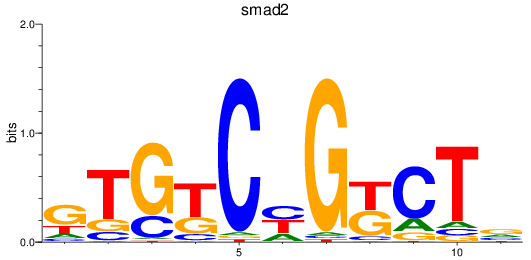
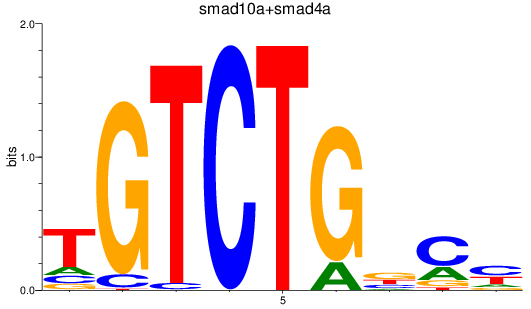
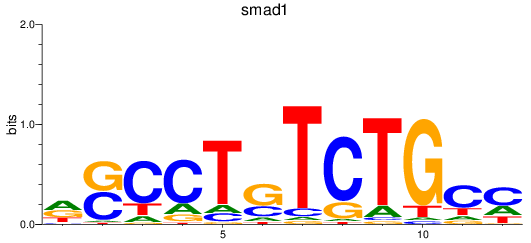
Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for smad2_smad10a+smad4a_smad1
Z-value: 0.75
Transcription factors associated with smad2_smad10a+smad4a_smad1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smad2
|
ENSDARG00000006389 | SMAD family member 2 |
|
smad10a
|
ENSDARG00000045094 | si_dkey-222b8.1 |
|
smad10a
|
ENSDARG00000070428 | si_dkey-222b8.1 |
|
smad4a
|
ENSDARG00000075226 | SMAD family member 4a |
|
smad1
|
ENSDARG00000027199 | SMAD family member 1 |
|
smad1
|
ENSDARG00000112617 | SMAD family member 1 |
|
smad1
|
ENSDARG00000115674 | SMAD family member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smad4a | dr11_v1_chr10_+_573667_573667 | 0.61 | 7.5e-03 | Click! |
| si:dkey-239n17.4 | dr11_v1_chr16_-_28876479_28876479 | -0.41 | 9.1e-02 | Click! |
| smad2 | dr11_v1_chr10_-_14929392_14929392 | 0.23 | 3.6e-01 | Click! |
| si:dkey-222b8.1 | dr11_v1_chr19_+_23932259_23932259 | 0.19 | 4.4e-01 | Click! |
| smad1 | dr11_v1_chr1_-_35916247_35916247 | -0.06 | 8.1e-01 | Click! |
Activity profile of smad2_smad10a+smad4a_smad1 motif
Sorted Z-values of smad2_smad10a+smad4a_smad1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_3960161 | 2.94 |
ENSDART00000111453
|
myo19
|
myosin XIX |
| chr6_-_31827597 | 2.67 |
ENSDART00000159400
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr10_-_10018794 | 2.47 |
ENSDART00000130734
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr10_+_20364009 | 2.05 |
ENSDART00000186139
ENSDART00000080395 |
golga7
|
golgin A7 |
| chr7_+_9290929 | 1.98 |
ENSDART00000128530
|
snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr24_-_26632171 | 1.98 |
ENSDART00000008374
ENSDART00000017384 |
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr2_+_54755172 | 1.61 |
ENSDART00000097864
|
ankrd12
|
ankyrin repeat domain 12 |
| chr16_+_40340222 | 1.57 |
ENSDART00000190631
|
mettl6
|
methyltransferase like 6 |
| chr10_+_37181780 | 1.53 |
ENSDART00000187625
|
ksr1a
|
kinase suppressor of ras 1a |
| chr13_+_31648271 | 1.44 |
ENSDART00000006648
|
mnat1
|
MNAT CDK-activating kinase assembly factor 1 |
| chr23_+_41831224 | 1.35 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr13_-_479129 | 1.23 |
ENSDART00000159803
ENSDART00000082127 |
heatr5b
|
HEAT repeat containing 5B |
| chr10_+_37182626 | 1.23 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr14_-_237130 | 1.22 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr18_-_25051846 | 1.22 |
ENSDART00000013082
|
st8sia2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr22_+_24559947 | 1.09 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr23_+_36460239 | 1.06 |
ENSDART00000172441
|
lima1a
|
LIM domain and actin binding 1a |
| chr5_+_3892551 | 1.05 |
ENSDART00000134396
|
rpain
|
RPA interacting protein |
| chr12_+_8822717 | 1.03 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr10_+_5744941 | 0.96 |
ENSDART00000159769
ENSDART00000184734 |
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr19_+_41551335 | 0.94 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr4_-_21466825 | 0.89 |
ENSDART00000066897
|
pawr
|
PRKC, apoptosis, WT1, regulator |
| chr19_-_5380770 | 0.87 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr20_-_2355357 | 0.77 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr22_-_506522 | 0.74 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr23_-_32334208 | 0.73 |
ENSDART00000053472
|
rnf41
|
ring finger protein 41 |
| chr1_+_57347888 | 0.71 |
ENSDART00000104222
|
b3gntl1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 |
| chr17_+_45765164 | 0.71 |
ENSDART00000145882
|
aspg
|
asparaginase homolog (S. cerevisiae) |
| chr18_+_27337994 | 0.70 |
ENSDART00000136172
|
si:dkey-29p10.4
|
si:dkey-29p10.4 |
| chr7_+_26998169 | 0.69 |
ENSDART00000128110
ENSDART00000101018 |
caprin1a
|
cell cycle associated protein 1a |
| chr13_+_15656042 | 0.67 |
ENSDART00000134240
|
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr8_-_30742233 | 0.66 |
ENSDART00000098986
|
gucd1
|
guanylyl cyclase domain containing 1 |
| chr9_+_44304980 | 0.65 |
ENSDART00000147990
|
ssfa2
|
sperm specific antigen 2 |
| chr20_-_4049862 | 0.64 |
ENSDART00000158057
|
sprtn
|
SprT-like N-terminal domain |
| chr25_+_418932 | 0.64 |
ENSDART00000059193
|
prtgb
|
protogenin homolog b (Gallus gallus) |
| chr15_+_11427620 | 0.62 |
ENSDART00000168688
|
ndufc2
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2 |
| chr10_+_44581378 | 0.62 |
ENSDART00000190331
|
sez6l
|
seizure related 6 homolog (mouse)-like |
| chr10_-_10018120 | 0.61 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr8_+_30742898 | 0.61 |
ENSDART00000018475
|
snrpd3
|
small nuclear ribonucleoprotein D3 polypeptide |
| chr7_-_48173440 | 0.60 |
ENSDART00000124075
|
mtss1lb
|
metastasis suppressor 1-like b |
| chr19_-_42503143 | 0.56 |
ENSDART00000007642
|
zgc:110239
|
zgc:110239 |
| chr19_-_3574060 | 0.56 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr13_-_1427240 | 0.56 |
ENSDART00000145867
|
znf451
|
zinc finger protein 451 |
| chr15_-_30984804 | 0.55 |
ENSDART00000157005
|
nf1a
|
neurofibromin 1a |
| chr2_-_37098785 | 0.55 |
ENSDART00000003670
|
zgc:101744
|
zgc:101744 |
| chr17_-_21280185 | 0.52 |
ENSDART00000123198
|
hspa12a
|
heat shock protein 12A |
| chr3_-_21137362 | 0.47 |
ENSDART00000104051
|
cdipt
|
CDP-diacylglycerol--inositol 3-phosphatidyltransferase (phosphatidylinositol synthase) |
| chr14_+_4276394 | 0.45 |
ENSDART00000038301
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr1_+_23372470 | 0.44 |
ENSDART00000187503
ENSDART00000087658 |
n4bp2
|
NEDD4 binding protein 2 |
| chr20_+_23625387 | 0.41 |
ENSDART00000147945
ENSDART00000150497 |
palld
|
palladin, cytoskeletal associated protein |
| chr2_-_57707039 | 0.41 |
ENSDART00000097685
|
CABZ01060891.1
|
|
| chr1_-_23294753 | 0.40 |
ENSDART00000013263
|
ugdh
|
UDP-glucose 6-dehydrogenase |
| chr12_+_33403694 | 0.40 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr2_-_55317035 | 0.38 |
ENSDART00000169382
ENSDART00000097874 |
tpm4b
|
tropomyosin 4b |
| chr21_+_33249478 | 0.36 |
ENSDART00000169972
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr23_-_46040618 | 0.36 |
ENSDART00000161415
|
CABZ01080918.1
|
|
| chr25_+_32496723 | 0.36 |
ENSDART00000087978
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr4_-_797831 | 0.36 |
ENSDART00000158970
ENSDART00000170012 |
mapre3b
|
microtubule-associated protein, RP/EB family, member 3b |
| chr15_-_47468085 | 0.36 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr11_-_44647286 | 0.35 |
ENSDART00000169329
ENSDART00000158939 |
tomm20b
|
translocase of outer mitochondrial membrane 20b |
| chr20_+_35085224 | 0.35 |
ENSDART00000040456
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr25_-_12805295 | 0.34 |
ENSDART00000157629
|
ca5a
|
carbonic anhydrase Va |
| chr18_+_50961953 | 0.34 |
ENSDART00000158768
|
ppfia1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 |
| chr9_+_12907574 | 0.34 |
ENSDART00000102348
|
si:dkey-230p4.1
|
si:dkey-230p4.1 |
| chr22_+_21549419 | 0.33 |
ENSDART00000139411
|
plpp2b
|
phospholipid phosphatase 2b |
| chr24_-_37272116 | 0.33 |
ENSDART00000022999
|
ube2ib
|
ubiquitin-conjugating enzyme E2Ib |
| chr18_+_3169579 | 0.33 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr14_-_6987649 | 0.33 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr25_+_32496877 | 0.32 |
ENSDART00000132698
|
ctdspl2a
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2a |
| chr8_-_14126646 | 0.32 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr19_-_18152942 | 0.31 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr17_+_43863708 | 0.30 |
ENSDART00000133874
ENSDART00000140316 ENSDART00000142929 ENSDART00000148090 |
zgc:66313
|
zgc:66313 |
| chr17_+_21902058 | 0.30 |
ENSDART00000142178
|
plekha1a
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1a |
| chr12_+_48681601 | 0.30 |
ENSDART00000187831
|
uros
|
uroporphyrinogen III synthase |
| chr25_-_27665978 | 0.28 |
ENSDART00000123590
|
si:ch211-91p5.3
|
si:ch211-91p5.3 |
| chr14_+_44805649 | 0.28 |
ENSDART00000180361
|
slc30a9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr8_+_19621511 | 0.28 |
ENSDART00000017128
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr11_-_44999858 | 0.28 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr23_-_41762956 | 0.27 |
ENSDART00000128302
|
stk35
|
serine/threonine kinase 35 |
| chr17_+_19626479 | 0.27 |
ENSDART00000044993
ENSDART00000131863 |
rgs7a
|
regulator of G protein signaling 7a |
| chr8_-_15520907 | 0.27 |
ENSDART00000133114
|
bend5
|
BEN domain containing 5 |
| chr2_+_47944967 | 0.27 |
ENSDART00000135584
|
ftr19
|
finTRIM family, member 19 |
| chr1_-_43905252 | 0.26 |
ENSDART00000135477
ENSDART00000132089 |
si:dkey-22i16.3
|
si:dkey-22i16.3 |
| chr13_-_15929402 | 0.26 |
ENSDART00000090273
|
ttl
|
tubulin tyrosine ligase |
| chr15_-_28904371 | 0.26 |
ENSDART00000155154
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr19_-_11425542 | 0.26 |
ENSDART00000177875
ENSDART00000080762 |
sept7a
|
septin 7a |
| chr2_-_49627099 | 0.26 |
ENSDART00000083710
ENSDART00000147421 |
cactin
|
cactin |
| chr5_-_70625697 | 0.26 |
ENSDART00000179538
ENSDART00000190540 |
CABZ01075572.1
|
|
| chr3_-_1317290 | 0.25 |
ENSDART00000047094
|
LO018552.1
|
|
| chr21_+_15883546 | 0.25 |
ENSDART00000186325
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr8_+_13064750 | 0.25 |
ENSDART00000039878
|
sap30bp
|
SAP30 binding protein |
| chr14_+_34558480 | 0.24 |
ENSDART00000075170
|
pttg1
|
pituitary tumor-transforming 1 |
| chr17_+_10748366 | 0.24 |
ENSDART00000018683
ENSDART00000097274 |
ATG14
|
zgc:113944 |
| chr23_-_24226533 | 0.24 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr23_-_29812667 | 0.23 |
ENSDART00000006120
|
pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
| chr10_-_35186310 | 0.23 |
ENSDART00000127805
|
pom121
|
POM121 transmembrane nucleoporin |
| chr13_+_22719789 | 0.23 |
ENSDART00000057672
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr19_-_81851 | 0.23 |
ENSDART00000172319
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_-_37785873 | 0.23 |
ENSDART00000011691
|
baxa
|
BCL2 associated X, apoptosis regulator a |
| chr2_+_26240339 | 0.22 |
ENSDART00000191006
|
palm1b
|
paralemmin 1b |
| chr20_-_31427390 | 0.22 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr23_-_32156278 | 0.22 |
ENSDART00000157479
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr16_-_30655980 | 0.22 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr3_-_15352303 | 0.21 |
ENSDART00000104338
ENSDART00000145919 ENSDART00000132135 |
pitpnbl
|
phosphatidylinositol transfer protein, beta, like |
| chr5_+_8964926 | 0.21 |
ENSDART00000091397
ENSDART00000164535 |
tnksb
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase b |
| chr21_-_9466201 | 0.21 |
ENSDART00000167211
|
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr12_+_48511605 | 0.21 |
ENSDART00000168389
|
arhgap44
|
Rho GTPase activating protein 44 |
| chr5_-_42180205 | 0.21 |
ENSDART00000145247
|
fam222ba
|
family with sequence similarity 222, member Ba |
| chr7_+_33424044 | 0.21 |
ENSDART00000180260
|
glceb
|
glucuronic acid epimerase b |
| chr12_-_5505205 | 0.20 |
ENSDART00000092319
|
abi3b
|
ABI family, member 3b |
| chr5_-_23517747 | 0.20 |
ENSDART00000137655
|
stag2a
|
stromal antigen 2a |
| chr5_-_58780160 | 0.20 |
ENSDART00000166955
|
arhgef12b
|
Rho guanine nucleotide exchange factor (GEF) 12b |
| chr20_+_46572550 | 0.20 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr7_-_26601307 | 0.20 |
ENSDART00000188934
|
plscr3b
|
phospholipid scramblase 3b |
| chr14_+_45608576 | 0.20 |
ENSDART00000173248
|
si:ch211-276i12.11
|
si:ch211-276i12.11 |
| chr10_+_1052591 | 0.20 |
ENSDART00000123405
|
unc5c
|
unc-5 netrin receptor C |
| chr7_+_16352924 | 0.20 |
ENSDART00000158972
|
mpped2a
|
metallophosphoesterase domain containing 2a |
| chr6_-_7107868 | 0.20 |
ENSDART00000171123
|
nhej1
|
nonhomologous end-joining factor 1 |
| chr25_+_3231812 | 0.20 |
ENSDART00000163647
|
metap2b
|
methionyl aminopeptidase 2b |
| chr9_-_32912638 | 0.20 |
ENSDART00000110582
|
fam160a2
|
family with sequence similarity 160, member A2 |
| chr3_-_56569968 | 0.20 |
ENSDART00000165317
|
SMIM10
|
Danio rerio uncharacterized LOC797202 (LOC797202), mRNA. |
| chr21_-_34032650 | 0.20 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr22_-_16270462 | 0.20 |
ENSDART00000105681
|
cdc14ab
|
cell division cycle 14Ab |
| chr23_+_9522942 | 0.19 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr15_-_30867594 | 0.19 |
ENSDART00000154448
|
nf1a
|
neurofibromin 1a |
| chr19_-_5058908 | 0.19 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr17_+_1514711 | 0.19 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr16_+_54641230 | 0.19 |
ENSDART00000157641
ENSDART00000159540 |
fbxo43
|
F-box protein 43 |
| chr19_+_41551543 | 0.19 |
ENSDART00000112364
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr6_+_44197348 | 0.19 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr19_-_15192638 | 0.19 |
ENSDART00000048151
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr8_-_30424182 | 0.19 |
ENSDART00000099021
|
dock8
|
dedicator of cytokinesis 8 |
| chr12_-_628024 | 0.19 |
ENSDART00000158563
ENSDART00000152612 |
wu:fj29h11
si:ch73-301j1.1
|
wu:fj29h11 si:ch73-301j1.1 |
| chr6_+_44197099 | 0.19 |
ENSDART00000124168
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr15_+_45595385 | 0.19 |
ENSDART00000161937
ENSDART00000170214 ENSDART00000157450 |
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr22_-_20419660 | 0.19 |
ENSDART00000105520
|
pias4a
|
protein inhibitor of activated STAT, 4a |
| chr23_-_21535040 | 0.19 |
ENSDART00000010647
|
rcc2
|
regulator of chromosome condensation 2 |
| chr15_+_17345609 | 0.18 |
ENSDART00000111753
|
vmp1
|
vacuole membrane protein 1 |
| chr11_+_21586335 | 0.18 |
ENSDART00000091182
|
FAM72B
|
zgc:101564 |
| chr20_-_2290529 | 0.18 |
ENSDART00000134695
|
EPB41L2
|
si:ch73-18b11.1 |
| chr23_+_30898013 | 0.18 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr23_+_9522781 | 0.18 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr1_-_50611031 | 0.18 |
ENSDART00000148285
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr12_+_19281189 | 0.18 |
ENSDART00000153428
|
tnrc6b
|
trinucleotide repeat containing 6b |
| chr2_+_8649293 | 0.18 |
ENSDART00000081442
ENSDART00000186024 |
fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr14_+_16036139 | 0.18 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr10_+_44956660 | 0.18 |
ENSDART00000169225
ENSDART00000189298 |
il1b
|
interleukin 1, beta |
| chr17_-_15546862 | 0.17 |
ENSDART00000091021
|
col10a1a
|
collagen, type X, alpha 1a |
| chr19_-_5058515 | 0.17 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr12_+_22628704 | 0.17 |
ENSDART00000168935
|
mfsd8
|
major facilitator superfamily domain containing 8 |
| chr4_-_5018705 | 0.17 |
ENSDART00000154025
|
strip2
|
striatin interacting protein 2 |
| chr14_+_22397251 | 0.17 |
ENSDART00000185239
ENSDART00000124072 ENSDART00000054977 |
atp7a
|
ATPase copper transporting alpha |
| chr23_+_12840080 | 0.17 |
ENSDART00000081016
ENSDART00000121697 |
smc1al
|
structural maintenance of chromosomes 1A, like |
| chr24_+_31334209 | 0.17 |
ENSDART00000168837
ENSDART00000172473 |
fam168b
|
family with sequence similarity 168, member B |
| chr7_-_60351876 | 0.17 |
ENSDART00000098563
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr24_-_16979728 | 0.17 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr20_-_1265562 | 0.17 |
ENSDART00000189866
|
lats1
|
large tumor suppressor kinase 1 |
| chr3_-_40051425 | 0.17 |
ENSDART00000146700
|
llgl1
|
lethal giant larvae homolog 1 (Drosophila) |
| chr14_+_50937757 | 0.17 |
ENSDART00000163865
|
rnf44
|
ring finger protein 44 |
| chr22_-_6420239 | 0.17 |
ENSDART00000148385
|
zgc:171699
|
zgc:171699 |
| chr8_+_10823069 | 0.17 |
ENSDART00000081341
|
mapk13
|
mitogen-activated protein kinase 13 |
| chr9_+_21793565 | 0.17 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr12_+_16953415 | 0.17 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr7_-_60351537 | 0.17 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr8_+_53311965 | 0.17 |
ENSDART00000130104
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr21_-_41369370 | 0.17 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr23_-_41762797 | 0.17 |
ENSDART00000186564
|
stk35
|
serine/threonine kinase 35 |
| chr9_-_29579908 | 0.17 |
ENSDART00000140876
|
cenpj
|
centromere protein J |
| chr20_-_34670236 | 0.16 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr15_-_30984557 | 0.16 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr25_-_21782435 | 0.16 |
ENSDART00000089616
|
bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr13_-_21650404 | 0.16 |
ENSDART00000078352
|
tspan14
|
tetraspanin 14 |
| chr7_-_5375214 | 0.16 |
ENSDART00000033316
|
vangl2
|
VANGL planar cell polarity protein 2 |
| chr21_+_43199237 | 0.16 |
ENSDART00000151748
|
aff4
|
AF4/FMR2 family, member 4 |
| chr24_-_26995164 | 0.16 |
ENSDART00000142864
|
stag1b
|
stromal antigen 1b |
| chr20_+_21268795 | 0.16 |
ENSDART00000090016
|
nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
| chr16_+_13822137 | 0.16 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr19_-_15192840 | 0.16 |
ENSDART00000151337
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr4_+_20051478 | 0.16 |
ENSDART00000143642
|
lamtor4
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chr5_+_40835601 | 0.16 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr11_-_23687158 | 0.16 |
ENSDART00000189599
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr17_+_20569806 | 0.16 |
ENSDART00000113936
|
SAMD8
|
zgc:162183 |
| chr7_-_26270014 | 0.16 |
ENSDART00000079347
|
serpine1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr7_-_19638319 | 0.16 |
ENSDART00000163686
|
si:ch211-212k18.4
|
si:ch211-212k18.4 |
| chr9_-_7213772 | 0.16 |
ENSDART00000174720
ENSDART00000092435 |
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr5_-_9678075 | 0.16 |
ENSDART00000097217
|
FAM109B
|
si:ch211-193c2.2 |
| chr12_-_3978306 | 0.16 |
ENSDART00000149473
ENSDART00000114857 |
ppp4cb
|
protein phosphatase 4, catalytic subunit b |
| chr21_-_41369539 | 0.16 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr2_+_44518636 | 0.16 |
ENSDART00000153733
|
pask
|
PAS domain containing serine/threonine kinase |
| chr25_+_22281441 | 0.16 |
ENSDART00000089516
|
stoml1
|
stomatin (EPB72)-like 1 |
| chr25_-_32363341 | 0.16 |
ENSDART00000153892
ENSDART00000114385 |
cep152
|
centrosomal protein 152 |
| chr7_+_24951552 | 0.16 |
ENSDART00000173644
|
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr23_-_27822920 | 0.15 |
ENSDART00000023094
|
acvr1ba
|
activin A receptor type 1Ba |
| chr12_+_33894665 | 0.15 |
ENSDART00000004769
|
mfsd13a
|
major facilitator superfamily domain containing 13A |
| chr17_+_10578823 | 0.15 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr3_-_29968015 | 0.15 |
ENSDART00000077119
ENSDART00000139310 |
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr17_+_23311377 | 0.15 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr5_-_21422390 | 0.15 |
ENSDART00000144198
|
tenm1
|
teneurin transmembrane protein 1 |
| chr7_+_17096281 | 0.15 |
ENSDART00000035558
|
htatip2
|
HIV-1 Tat interactive protein 2 |
| chr2_-_2642476 | 0.15 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of smad2_smad10a+smad4a_smad1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.3 | 1.3 | GO:0032370 | positive regulation of lipid transport(GO:0032370) |
| 0.2 | 1.0 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 0.6 | GO:0071788 | endoplasmic reticulum tubular network maintenance(GO:0071788) |
| 0.2 | 1.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 0.9 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.4 | GO:0006041 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 1.1 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 2.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.8 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.1 | 0.3 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.1 | 0.3 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.1 | 0.7 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.3 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.1 | 0.3 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) |
| 0.1 | 0.4 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.1 | 1.2 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.1 | 0.4 | GO:0003188 | heart valve formation(GO:0003188) atrioventricular valve formation(GO:0003190) |
| 0.1 | 0.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.1 | 0.2 | GO:1904353 | positive regulation of telomere maintenance(GO:0032206) regulation of telomere capping(GO:1904353) positive regulation of telomere capping(GO:1904355) |
| 0.1 | 1.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 2.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.3 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.6 | GO:2000142 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.3 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.4 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.2 | GO:0010755 | regulation of plasminogen activation(GO:0010755) |
| 0.0 | 0.1 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.0 | 0.3 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.2 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 0.2 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.1 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.0 | 0.2 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.2 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.1 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:0008591 | regulation of Wnt signaling pathway, calcium modulating pathway(GO:0008591) |
| 0.0 | 0.0 | GO:0014066 | regulation of phosphatidylinositol 3-kinase signaling(GO:0014066) |
| 0.0 | 0.1 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.2 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.3 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.6 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.0 | 0.1 | GO:2001014 | skeletal muscle fiber differentiation(GO:0098528) regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.0 | 0.1 | GO:0006266 | DNA ligation(GO:0006266) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.2 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.4 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 2.9 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.2 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0001836 | release of cytochrome c from mitochondria(GO:0001836) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.3 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.1 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0006178 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine biosynthetic process(GO:0046099) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.2 | GO:0046620 | secondary heart field specification(GO:0003139) regulation of organ growth(GO:0046620) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 1.2 | GO:0051170 | nuclear import(GO:0051170) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.1 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.1 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 0.1 | GO:0014015 | positive regulation of gliogenesis(GO:0014015) positive regulation of glial cell differentiation(GO:0045687) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.1 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) |
| 0.0 | 0.1 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.0 | 0.1 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.0 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0055117 | regulation of cardiac muscle contraction(GO:0055117) |
| 0.0 | 0.0 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.2 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.2 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.1 | GO:0035989 | tendon development(GO:0035989) |
| 0.0 | 0.2 | GO:0098754 | detoxification(GO:0098754) |
| 0.0 | 0.1 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.2 | GO:0033048 | negative regulation of sister chromatid segregation(GO:0033046) negative regulation of mitotic sister chromatid segregation(GO:0033048) negative regulation of chromosome segregation(GO:0051985) negative regulation of mitotic sister chromatid separation(GO:2000816) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.1 | 1.4 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.1 | 0.4 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 2.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.3 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.3 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0005775 | vacuolar lumen(GO:0005775) lysosomal lumen(GO:0043202) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0008352 | katanin complex(GO:0008352) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 1.0 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.0 | 2.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.1 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 1.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.0 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.2 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.2 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.3 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 1.0 | GO:0005882 | intermediate filament(GO:0005882) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.0 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.2 | 1.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.2 | 1.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 0.4 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 1.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.4 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.1 | 0.7 | GO:0004067 | asparaginase activity(GO:0004067) |
| 0.1 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.1 | 1.2 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 2.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.3 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 0.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.8 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 3.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 2.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 2.0 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.1 | GO:0033897 | ribonuclease T2 activity(GO:0033897) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.0 | 0.5 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.0 | 0.1 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.0 | 0.1 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.1 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.0 | 0.2 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0032357 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.3 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.2 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.1 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.4 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.0 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.4 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.2 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.9 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.3 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.1 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 1.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME EARLY PHASE OF HIV LIFE CYCLE | Genes involved in Early Phase of HIV Life Cycle |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.3 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |