Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
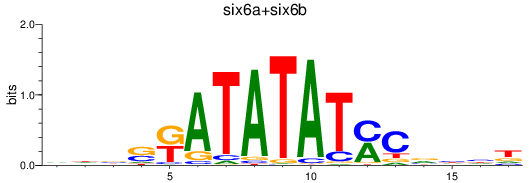
Results for six6a+six6b
Z-value: 0.67
Transcription factors associated with six6a+six6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six6a
|
ENSDARG00000025187 | SIX homeobox 6a |
|
six6b
|
ENSDARG00000031316 | SIX homeobox 6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six6b | dr11_v1_chr20_-_20533865_20533865 | -0.78 | 1.2e-04 | Click! |
| six6a | dr11_v1_chr13_+_31583034_31583034 | 0.49 | 3.7e-02 | Click! |
Activity profile of six6a+six6b motif
Sorted Z-values of six6a+six6b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_38883388 | 1.70 |
ENSDART00000135902
|
map2
|
microtubule-associated protein 2 |
| chr6_-_23002373 | 1.68 |
ENSDART00000037709
ENSDART00000170369 |
nol11
|
nucleolar protein 11 |
| chr24_-_26632171 | 1.56 |
ENSDART00000008374
ENSDART00000017384 |
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr2_-_6065416 | 1.37 |
ENSDART00000037698
|
uck2b
|
uridine-cytidine kinase 2b |
| chr15_+_46313082 | 1.26 |
ENSDART00000153830
|
si:ch1073-190k2.1
|
si:ch1073-190k2.1 |
| chr8_+_17143501 | 1.24 |
ENSDART00000061758
|
mier3b
|
mesoderm induction early response 1, family member 3 b |
| chr5_-_43859148 | 1.23 |
ENSDART00000162746
ENSDART00000128763 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr4_+_23117557 | 1.21 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr13_+_46944607 | 1.20 |
ENSDART00000187352
|
fbxo5
|
F-box protein 5 |
| chr2_-_23391266 | 1.16 |
ENSDART00000159048
|
ivns1abpb
|
influenza virus NS1A binding protein b |
| chr10_+_39199547 | 1.15 |
ENSDART00000075943
|
ei24
|
etoposide induced 2.4 |
| chr10_+_23060391 | 1.15 |
ENSDART00000079711
|
slc25a1a
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1a |
| chr7_+_71955486 | 1.14 |
ENSDART00000189349
|
CABZ01071171.1
|
Danio rerio low density lipoprotein receptor-related protein 4 (lrp4), mRNA. |
| chr8_-_12847483 | 1.14 |
ENSDART00000146186
|
si:dkey-104n9.1
|
si:dkey-104n9.1 |
| chr7_+_22313533 | 1.14 |
ENSDART00000123457
|
TMEM102
|
si:dkey-11f12.2 |
| chr11_+_1867613 | 1.11 |
ENSDART00000065470
|
rbms2a
|
RNA binding motif, single stranded interacting protein 2a |
| chr13_+_49727333 | 1.05 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr24_-_19719240 | 1.04 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr8_+_23826985 | 1.02 |
ENSDART00000187430
|
mapk14a
|
mitogen-activated protein kinase 14a |
| chr1_+_31657842 | 1.02 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr2_+_1881334 | 1.01 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr17_+_28706946 | 0.99 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr1_+_31658011 | 0.97 |
ENSDART00000192203
|
poll
|
polymerase (DNA directed), lambda |
| chr5_+_36661058 | 0.96 |
ENSDART00000125653
|
capns1a
|
calpain, small subunit 1 a |
| chr16_+_30002605 | 0.96 |
ENSDART00000160555
|
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr11_+_45286911 | 0.94 |
ENSDART00000181763
|
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr9_-_11676491 | 0.91 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr25_+_1335530 | 0.90 |
ENSDART00000090803
|
fem1b
|
fem-1 homolog b (C. elegans) |
| chr2_+_30249977 | 0.90 |
ENSDART00000109160
ENSDART00000135171 |
tmem70
|
transmembrane protein 70 |
| chr19_+_41479990 | 0.89 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr19_+_206835 | 0.85 |
ENSDART00000161137
|
scn1bb
|
sodium channel, voltage-gated, type I, beta b |
| chr23_-_36439961 | 0.85 |
ENSDART00000187907
|
csad
|
cysteine sulfinic acid decarboxylase |
| chr12_-_27212596 | 0.84 |
ENSDART00000153101
|
psme3
|
proteasome activator subunit 3 |
| chr7_+_38808027 | 0.82 |
ENSDART00000052323
|
harbi1
|
harbinger transposase derived 1 |
| chr3_+_13850163 | 0.81 |
ENSDART00000164872
|
ilf3b
|
interleukin enhancer binding factor 3b |
| chr15_-_25094026 | 0.81 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr23_+_1702624 | 0.80 |
ENSDART00000149357
|
rabggta
|
Rab geranylgeranyltransferase, alpha subunit |
| chr9_-_53062083 | 0.80 |
ENSDART00000122155
|
zmp:0000000936
|
zmp:0000000936 |
| chr5_+_25304499 | 0.80 |
ENSDART00000163425
|
carnmt1
|
carnosine N-methyltransferase 1 |
| chr5_-_39736383 | 0.79 |
ENSDART00000127123
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr25_+_33072676 | 0.78 |
ENSDART00000182885
ENSDART00000181516 |
tln2b
|
talin 2b |
| chr19_-_46018152 | 0.77 |
ENSDART00000159206
|
krit1
|
KRIT1, ankyrin repeat containing |
| chr1_+_17527342 | 0.77 |
ENSDART00000139702
ENSDART00000140076 ENSDART00000005593 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr15_-_34934784 | 0.76 |
ENSDART00000190848
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr17_+_25331576 | 0.76 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr12_+_47448558 | 0.76 |
ENSDART00000185689
ENSDART00000105328 |
fmn2b
|
formin 2b |
| chr14_+_22467672 | 0.75 |
ENSDART00000079409
ENSDART00000136597 |
nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr4_-_75172216 | 0.74 |
ENSDART00000127522
|
naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae) |
| chr23_-_36934944 | 0.71 |
ENSDART00000109976
ENSDART00000162179 |
acap3a
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3a |
| chr10_-_38243851 | 0.68 |
ENSDART00000166108
|
usp25
|
ubiquitin specific peptidase 25 |
| chr8_-_2602572 | 0.68 |
ENSDART00000110482
|
zdhhc12a
|
zinc finger, DHHC-type containing 12a |
| chr18_+_16133595 | 0.64 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr1_+_45925365 | 0.64 |
ENSDART00000144245
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr2_+_37245382 | 0.63 |
ENSDART00000004626
|
sec62
|
SEC62 homolog, preprotein translocation factor |
| chr14_+_26759332 | 0.62 |
ENSDART00000088484
|
ahnak
|
AHNAK nucleoprotein |
| chr8_-_38810233 | 0.61 |
ENSDART00000085304
|
pcsk5b
|
proprotein convertase subtilisin/kexin type 5b |
| chr15_-_34878388 | 0.59 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr13_-_24260609 | 0.59 |
ENSDART00000138747
|
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr2_-_9607879 | 0.58 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr15_-_25527580 | 0.57 |
ENSDART00000167005
ENSDART00000157498 |
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr3_-_12227359 | 0.57 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr2_+_51796441 | 0.56 |
ENSDART00000165151
|
crygn1
|
crystallin, gamma N1 |
| chr15_-_37867995 | 0.55 |
ENSDART00000192698
|
si:dkey-238d18.4
|
si:dkey-238d18.4 |
| chr3_-_21106093 | 0.54 |
ENSDART00000156566
|
maza
|
MYC-associated zinc finger protein a (purine-binding transcription factor) |
| chr10_+_44903676 | 0.52 |
ENSDART00000158553
|
zgc:114173
|
zgc:114173 |
| chr18_+_38192499 | 0.51 |
ENSDART00000191849
|
nucb2b
|
nucleobindin 2b |
| chr21_+_6197223 | 0.50 |
ENSDART00000147716
|
si:dkey-93m18.3
|
si:dkey-93m18.3 |
| chr19_-_20777351 | 0.49 |
ENSDART00000019206
|
ngly1
|
N-glycanase 1 |
| chr5_-_66749535 | 0.46 |
ENSDART00000132183
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr5_-_41638039 | 0.46 |
ENSDART00000144525
|
epg5
|
ectopic P-granules autophagy protein 5 homolog (C. elegans) |
| chr2_+_23039041 | 0.46 |
ENSDART00000056921
|
csnk1g2a
|
casein kinase 1, gamma 2a |
| chr23_+_16831063 | 0.45 |
ENSDART00000104782
|
zgc:173570
|
zgc:173570 |
| chr16_-_36979592 | 0.45 |
ENSDART00000168443
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr20_-_51656512 | 0.45 |
ENSDART00000129965
|
LO018154.1
|
|
| chr7_+_24006875 | 0.43 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr1_-_55888970 | 0.42 |
ENSDART00000064194
|
asf1bb
|
anti-silencing function 1Bb histone chaperone |
| chr6_-_9646275 | 0.42 |
ENSDART00000012903
|
wdr12
|
WD repeat domain 12 |
| chr24_-_7587401 | 0.41 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr14_+_50770537 | 0.40 |
ENSDART00000158723
|
sncb
|
synuclein, beta |
| chr3_-_39305291 | 0.38 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr10_+_45302425 | 0.38 |
ENSDART00000159954
|
zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr20_+_42918755 | 0.35 |
ENSDART00000134855
|
efr3bb
|
EFR3 homolog Bb (S. cerevisiae) |
| chr7_+_29177191 | 0.34 |
ENSDART00000008096
|
aph1b
|
APH1B gamma secretase subunit |
| chr18_-_41164277 | 0.34 |
ENSDART00000187766
ENSDART00000185375 |
CABZ01005876.1
|
|
| chr18_+_43183749 | 0.34 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr23_+_7710447 | 0.33 |
ENSDART00000168199
|
kif3b
|
kinesin family member 3B |
| chr8_+_10001805 | 0.33 |
ENSDART00000132894
|
si:dkey-8e10.2
|
si:dkey-8e10.2 |
| chr20_-_36393555 | 0.33 |
ENSDART00000153421
|
si:dkey-1j5.4
|
si:dkey-1j5.4 |
| chr22_-_21845685 | 0.33 |
ENSDART00000105564
|
aes
|
amino-terminal enhancer of split |
| chr6_-_16456093 | 0.31 |
ENSDART00000083305
ENSDART00000181640 |
slc19a2
|
solute carrier family 19 (thiamine transporter), member 2 |
| chr8_-_8346023 | 0.31 |
ENSDART00000164731
ENSDART00000188259 |
tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae) |
| chr7_+_69428831 | 0.31 |
ENSDART00000186975
|
elp5
|
elongator acetyltransferase complex subunit 5 |
| chr20_+_26987416 | 0.31 |
ENSDART00000012816
|
sel1l
|
sel-1 suppressor of lin-12-like (C. elegans) |
| chr23_+_30898013 | 0.31 |
ENSDART00000146859
|
cables2a
|
Cdk5 and Abl enzyme substrate 2a |
| chr19_-_32710922 | 0.30 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr11_+_5681762 | 0.30 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr6_-_50685862 | 0.29 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr11_-_27702778 | 0.29 |
ENSDART00000045942
ENSDART00000125352 |
phf2
|
PHD finger protein 2 |
| chr18_+_12058403 | 0.28 |
ENSDART00000140854
ENSDART00000193632 ENSDART00000190519 ENSDART00000190685 ENSDART00000112671 |
bicd1a
|
bicaudal D homolog 1a |
| chr10_-_24648228 | 0.28 |
ENSDART00000081834
ENSDART00000132830 |
stoml3b
|
stomatin (EPB72)-like 3b |
| chr11_-_17755444 | 0.28 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr13_+_22555342 | 0.27 |
ENSDART00000193633
|
bmpr1aa
|
bone morphogenetic protein receptor, type IAa |
| chr2_+_13907452 | 0.27 |
ENSDART00000169724
ENSDART00000190691 |
zgc:66475
|
zgc:66475 |
| chr5_-_57526807 | 0.27 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr25_-_10630496 | 0.27 |
ENSDART00000153639
ENSDART00000181722 ENSDART00000177834 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr3_+_18840810 | 0.27 |
ENSDART00000181137
ENSDART00000128626 ENSDART00000133332 |
tmem104
|
transmembrane protein 104 |
| chr15_-_31419805 | 0.27 |
ENSDART00000060111
|
or111-11
|
odorant receptor, family D, subfamily 111, member 11 |
| chr15_-_6946286 | 0.27 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr22_-_12862415 | 0.26 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr15_+_34934568 | 0.26 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr1_-_19233890 | 0.25 |
ENSDART00000127145
|
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr19_-_18152942 | 0.24 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr18_+_36786842 | 0.24 |
ENSDART00000123264
|
si:ch211-160d20.3
|
si:ch211-160d20.3 |
| chr8_-_28357177 | 0.23 |
ENSDART00000182319
|
KLHL12 (1 of many)
|
kelch like family member 12 |
| chr20_-_51946052 | 0.23 |
ENSDART00000074325
|
dusp10
|
dual specificity phosphatase 10 |
| chr25_+_15287036 | 0.23 |
ENSDART00000147572
|
hipk3a
|
homeodomain interacting protein kinase 3a |
| chr24_+_13017586 | 0.23 |
ENSDART00000142457
|
stau2
|
staufen double-stranded RNA binding protein 2 |
| chr22_+_19640309 | 0.22 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr5_+_37890521 | 0.22 |
ENSDART00000140207
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr4_+_76426308 | 0.22 |
ENSDART00000180424
ENSDART00000159789 |
FP074874.1
zgc:172128
|
zgc:172128 |
| chr15_+_11825366 | 0.20 |
ENSDART00000190042
|
prkd2
|
protein kinase D2 |
| chr8_-_21142550 | 0.20 |
ENSDART00000143192
ENSDART00000186820 ENSDART00000135938 |
cpt2
|
carnitine palmitoyltransferase 2 |
| chr10_-_43540196 | 0.19 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr6_-_49159207 | 0.19 |
ENSDART00000041942
|
tspan2a
|
tetraspanin 2a |
| chr15_-_762319 | 0.19 |
ENSDART00000154306
ENSDART00000157492 |
si:dkey-7i4.16
znf1011
|
si:dkey-7i4.16 zinc finger protein 1011 |
| chr7_-_36358735 | 0.19 |
ENSDART00000188392
|
fto
|
fat mass and obesity associated |
| chr7_-_7766920 | 0.18 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr1_+_34701144 | 0.18 |
ENSDART00000150038
|
gtf2f2a
|
general transcription factor IIF, polypeptide 2a |
| chr2_+_53357953 | 0.18 |
ENSDART00000187577
|
celf5b
|
cugbp, Elav-like family member 5b |
| chr12_-_43428542 | 0.17 |
ENSDART00000192266
|
ptprea
|
protein tyrosine phosphatase, receptor type, E, a |
| chr24_-_25788841 | 0.15 |
ENSDART00000132235
|
klhl24b
|
kelch-like family member 24b |
| chr10_-_25816558 | 0.15 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr5_+_18047111 | 0.14 |
ENSDART00000132164
|
hira
|
histone cell cycle regulator a |
| chr21_+_51521 | 0.14 |
ENSDART00000007672
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr3_+_41585298 | 0.14 |
ENSDART00000182370
|
card11
|
caspase recruitment domain family, member 11 |
| chr20_-_29471577 | 0.13 |
ENSDART00000043382
|
grem1a
|
gremlin 1a, DAN family BMP antagonist |
| chr9_-_22310919 | 0.13 |
ENSDART00000108719
|
crygm2d10
|
crystallin, gamma M2d10 |
| chr7_+_6385664 | 0.13 |
ENSDART00000173144
|
si:ch73-368j24.17
|
si:ch73-368j24.17 |
| chr23_+_41679586 | 0.12 |
ENSDART00000067662
|
CU914487.1
|
|
| chr5_-_38820046 | 0.11 |
ENSDART00000182886
|
cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr2_-_732864 | 0.10 |
ENSDART00000168585
|
CU861459.1
|
|
| chr14_-_17306261 | 0.10 |
ENSDART00000191747
|
jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr21_-_1644414 | 0.09 |
ENSDART00000105736
ENSDART00000124904 |
zgc:152948
|
zgc:152948 |
| chr1_-_45614318 | 0.09 |
ENSDART00000149725
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr4_+_76752707 | 0.08 |
ENSDART00000082121
|
ms4a17a.4
|
membrane-spanning 4-domains, subfamily A, member 17A.4 |
| chr11_+_44804685 | 0.08 |
ENSDART00000163660
|
strn
|
striatin, calmodulin binding protein |
| chr17_+_41081512 | 0.08 |
ENSDART00000056742
|
babam2
|
BRISC and BRCA1 A complex member 2 |
| chr2_-_32366989 | 0.07 |
ENSDART00000136758
|
ubtfl
|
upstream binding transcription factor, like |
| chr3_-_61387273 | 0.07 |
ENSDART00000156479
|
znf1143
|
zinc finger protein 1143 |
| chr16_+_3185541 | 0.07 |
ENSDART00000024088
|
wdr21
|
WD repeat domain 21 |
| chr22_+_2844865 | 0.06 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr10_-_36225729 | 0.06 |
ENSDART00000127677
|
or109-5
|
odorant receptor, family D, subfamily 109, member 5 |
| chr22_+_9751117 | 0.04 |
ENSDART00000121827
|
BX664625.1
|
|
| chr14_-_736575 | 0.04 |
ENSDART00000168611
|
tlr1
|
toll-like receptor 1 |
| chr20_-_37287107 | 0.04 |
ENSDART00000076309
|
nmbr
|
neuromedin B receptor |
| chr24_-_20016817 | 0.04 |
ENSDART00000082201
ENSDART00000189448 |
slc22a13b
|
solute carrier family 22 member 13b |
| chr20_-_16849306 | 0.04 |
ENSDART00000131395
ENSDART00000027582 |
brms1lb
|
breast cancer metastasis-suppressor 1-like b |
| chr23_-_38497705 | 0.03 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr17_-_15611744 | 0.03 |
ENSDART00000010496
|
fhl5
|
four and a half LIM domains 5 |
| chr20_+_1960092 | 0.03 |
ENSDART00000191892
|
CABZ01103860.1
|
|
| chr4_-_41712014 | 0.03 |
ENSDART00000138165
|
znf976
|
zinc finger protein 976 |
| chr1_+_23408622 | 0.03 |
ENSDART00000140706
|
chrna9
|
cholinergic receptor, nicotinic, alpha 9 |
| chr11_+_2398843 | 0.03 |
ENSDART00000126761
|
pip4k2cb
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma b |
| chr20_+_5132138 | 0.03 |
ENSDART00000081351
ENSDART00000157685 |
cyp46a1.1
|
cytochrome P450, family 46, subfamily A, polypeptide 1, tandem duplicate 1 |
| chr18_+_32979166 | 0.02 |
ENSDART00000163528
|
olfcg4
|
olfactory receptor C family, g4 |
| chr6_-_28980756 | 0.02 |
ENSDART00000014661
|
glmnb
|
glomulin, FKBP associated protein b |
| chr22_-_10459880 | 0.02 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr23_+_32335871 | 0.01 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr9_+_50007556 | 0.01 |
ENSDART00000175587
|
slc38a11
|
solute carrier family 38, member 11 |
| chr7_-_44970682 | 0.01 |
ENSDART00000144591
|
fam96b
|
family with sequence similarity 96, member B |
| chr9_+_41156818 | 0.01 |
ENSDART00000105764
ENSDART00000147052 |
stat4
|
signal transducer and activator of transcription 4 |
| chr2_+_3881000 | 0.01 |
ENSDART00000081897
|
mpp7b
|
membrane protein, palmitoylated 7b (MAGUK p55 subfamily member 7) |
| chr8_-_8698607 | 0.01 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr18_-_19350792 | 0.01 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr25_-_24248000 | 0.00 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr6_-_18199062 | 0.00 |
ENSDART00000167513
|
ppp1r27b
|
protein phosphatase 1, regulatory subunit 27b |
| chr15_-_669476 | 0.00 |
ENSDART00000153687
ENSDART00000030603 |
si:ch211-210b2.2
|
si:ch211-210b2.2 |
| chr11_-_34166960 | 0.00 |
ENSDART00000181571
|
atp13a3
|
ATPase 13A3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of six6a+six6b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:1901836 | regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901836) |
| 0.3 | 1.2 | GO:0051444 | negative regulation of ligase activity(GO:0051352) negative regulation of ubiquitin-protein transferase activity(GO:0051444) negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.3 | 0.9 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.3 | 1.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.3 | 0.8 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) regulation of endothelial cell differentiation(GO:0045601) |
| 0.3 | 0.8 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 0.9 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.2 | 1.4 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 1.0 | GO:1901741 | embryonic cleavage(GO:0040016) positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.9 | GO:0086091 | regulation of heart rate by cardiac conduction(GO:0086091) |
| 0.1 | 0.6 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.1 | 0.8 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0019062 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.3 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.1 | 0.6 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 1.5 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.1 | 2.0 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.7 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.5 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.3 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.6 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.5 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) glutamine catabolic process(GO:0006543) |
| 0.0 | 0.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0045117 | azole transport(GO:0045117) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.4 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 1.1 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.4 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 1.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.8 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 1.0 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.7 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.4 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.8 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 0.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.9 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.3 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 0.3 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.6 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.0 | 0.5 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.8 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.4 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.2 | 0.9 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 0.8 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.2 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.7 | GO:0043998 | H2A histone acetyltransferase activity(GO:0043998) |
| 0.2 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.4 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 0.8 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.6 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.6 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.8 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 0.4 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.1 | 0.9 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 2.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.8 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 1.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 1.2 | GO:0005338 | nucleotide-sugar transmembrane transporter activity(GO:0005338) |
| 0.1 | 1.1 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.2 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.3 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.3 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.8 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.2 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.7 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.1 | 2.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.8 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 1.2 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.8 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.3 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |