Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
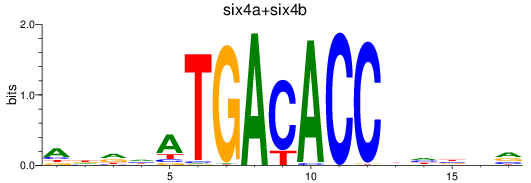
Results for six4a+six4b
Z-value: 0.73
Transcription factors associated with six4a+six4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six4a
|
ENSDARG00000004695 | SIX homeobox 4a |
|
six4b
|
ENSDARG00000031983 | SIX homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six4b | dr11_v1_chr20_+_20484827_20484827 | -0.85 | 7.5e-06 | Click! |
| six4a | dr11_v1_chr13_-_31647323_31647323 | -0.71 | 1.1e-03 | Click! |
Activity profile of six4a+six4b motif
Sorted Z-values of six4a+six4b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_25236657 | 2.33 |
ENSDART00000138012
|
zgc:172218
|
zgc:172218 |
| chr2_+_68789 | 2.31 |
ENSDART00000058569
|
cldn1
|
claudin 1 |
| chr22_+_25242322 | 2.14 |
ENSDART00000134628
|
si:ch211-226h8.8
|
si:ch211-226h8.8 |
| chr22_+_25236888 | 1.99 |
ENSDART00000037286
|
zgc:172218
|
zgc:172218 |
| chr3_-_54607166 | 1.64 |
ENSDART00000021977
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr9_+_2041535 | 1.62 |
ENSDART00000093187
|
lnpa
|
limb and neural patterns a |
| chr5_+_13472234 | 1.58 |
ENSDART00000114069
ENSDART00000132406 |
cnnm4b
|
cyclin and CBS domain divalent metal cation transport mediator 4b |
| chr9_+_8396755 | 1.31 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr15_-_35126332 | 1.29 |
ENSDART00000007636
|
zgc:55413
|
zgc:55413 |
| chr18_+_27511976 | 1.17 |
ENSDART00000132017
ENSDART00000140781 |
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr1_-_6085750 | 1.12 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr19_-_11031145 | 1.09 |
ENSDART00000151375
ENSDART00000027598 ENSDART00000137865 ENSDART00000188025 |
tpm3
|
tropomyosin 3 |
| chr19_-_6983002 | 1.02 |
ENSDART00000104891
|
znf384l
|
zinc finger protein 384 like |
| chr16_-_17347727 | 1.01 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr12_+_1592146 | 1.01 |
ENSDART00000184575
ENSDART00000192902 |
SLC39A11
|
solute carrier family 39 member 11 |
| chr7_-_60351537 | 1.00 |
ENSDART00000159875
|
plcb3
|
phospholipase C, beta 3 (phosphatidylinositol-specific) |
| chr20_+_26943072 | 0.93 |
ENSDART00000153215
|
cdca4
|
cell division cycle associated 4 |
| chr16_+_35905031 | 0.87 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr7_+_48297842 | 0.81 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr20_-_51831657 | 0.80 |
ENSDART00000165076
|
mia3
|
melanoma inhibitory activity family, member 3 |
| chr6_-_3978919 | 0.75 |
ENSDART00000167753
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr9_+_27750490 | 0.75 |
ENSDART00000125303
ENSDART00000189279 ENSDART00000021065 ENSDART00000148060 |
rab5b
|
RAB5B, member RAS oncogene family |
| chr22_+_2751887 | 0.75 |
ENSDART00000133652
|
si:dkey-20i20.11
|
si:dkey-20i20.11 |
| chr12_-_4475890 | 0.72 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr7_-_58130703 | 0.72 |
ENSDART00000172082
|
ank2b
|
ankyrin 2b, neuronal |
| chr24_-_23839647 | 0.72 |
ENSDART00000125190
|
rrs1
|
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
| chr10_+_39283985 | 0.71 |
ENSDART00000016464
|
dcps
|
decapping enzyme, scavenger |
| chr19_-_6083761 | 0.70 |
ENSDART00000151185
ENSDART00000143941 |
gsk3aa
|
glycogen synthase kinase 3 alpha a |
| chr2_-_43739559 | 0.68 |
ENSDART00000138947
|
kif5ba
|
kinesin family member 5B, a |
| chr3_+_52953489 | 0.67 |
ENSDART00000125136
|
dsn1
|
DSN1 homolog, MIS12 kinetochore complex component |
| chr17_-_13072334 | 0.66 |
ENSDART00000159598
|
CU469462.1
|
|
| chr2_-_40135942 | 0.64 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr16_+_50755133 | 0.64 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr1_+_37752171 | 0.61 |
ENSDART00000183247
ENSDART00000189756 ENSDART00000139448 |
GALNTL6
|
si:ch211-15e22.3 |
| chr18_-_39188664 | 0.60 |
ENSDART00000162983
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr3_+_18579806 | 0.60 |
ENSDART00000180967
ENSDART00000089765 |
arhgap17b
|
Rho GTPase activating protein 17b |
| chr7_-_54217547 | 0.58 |
ENSDART00000162777
ENSDART00000188268 ENSDART00000165875 |
csnk1g1
|
casein kinase 1, gamma 1 |
| chr20_+_37866686 | 0.58 |
ENSDART00000036546
|
vash2
|
vasohibin 2 |
| chr10_-_39283883 | 0.58 |
ENSDART00000023831
|
cry5
|
cryptochrome circadian clock 5 |
| chr8_+_35964482 | 0.57 |
ENSDART00000129357
ENSDART00000154953 |
glt1d1
|
glycosyltransferase 1 domain containing 1 |
| chr2_-_30770736 | 0.56 |
ENSDART00000131230
|
rgs20
|
regulator of G protein signaling 20 |
| chr23_+_27779452 | 0.55 |
ENSDART00000134785
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr21_+_34167178 | 0.49 |
ENSDART00000158308
|
trpc5b
|
transient receptor potential cation channel, subfamily C, member 5b |
| chr12_+_23991276 | 0.48 |
ENSDART00000153136
|
psme4b
|
proteasome activator subunit 4b |
| chr20_-_6184167 | 0.48 |
ENSDART00000147451
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr25_+_5604512 | 0.48 |
ENSDART00000042781
|
plxnb2b
|
plexin b2b |
| chr11_+_24851671 | 0.45 |
ENSDART00000167659
|
ipo9
|
importin 9 |
| chr7_+_38770167 | 0.45 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr24_-_25166416 | 0.45 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr6_-_40657653 | 0.42 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr2_-_20666920 | 0.39 |
ENSDART00000143437
ENSDART00000114546 ENSDART00000136113 ENSDART00000179247 |
dusp12
|
dual specificity phosphatase 12 |
| chr4_+_288633 | 0.39 |
ENSDART00000183304
|
FO834823.1
|
|
| chr9_-_34882516 | 0.38 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr5_+_12908781 | 0.38 |
ENSDART00000051663
|
ppil2
|
peptidylprolyl isomerase (cyclophilin)-like 2 |
| chr5_+_66044128 | 0.38 |
ENSDART00000165573
|
jak2b
|
Janus kinase 2b |
| chr12_-_20658285 | 0.37 |
ENSDART00000080888
|
snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr11_+_1602916 | 0.35 |
ENSDART00000184434
ENSDART00000112597 ENSDART00000192165 |
si:dkey-40c23.2
si:dkey-40c23.3
|
si:dkey-40c23.2 si:dkey-40c23.3 |
| chr21_+_34686764 | 0.33 |
ENSDART00000005479
|
chm
|
choroideremia (Rab escort protein 1) |
| chr20_+_22067337 | 0.33 |
ENSDART00000152636
|
clocka
|
clock circadian regulator a |
| chr3_+_15773991 | 0.32 |
ENSDART00000089923
|
znf652
|
zinc finger protein 652 |
| chr10_-_32610776 | 0.32 |
ENSDART00000017436
|
mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr2_-_30784198 | 0.31 |
ENSDART00000182523
ENSDART00000147355 |
rgs20
|
regulator of G protein signaling 20 |
| chr13_+_4883377 | 0.31 |
ENSDART00000181530
ENSDART00000092521 |
micu1
|
mitochondrial calcium uptake 1 |
| chr5_+_55221593 | 0.30 |
ENSDART00000073638
|
tmc2a
|
transmembrane channel-like 2a |
| chr23_+_45785563 | 0.30 |
ENSDART00000186027
|
CABZ01088036.1
|
|
| chr19_-_10330778 | 0.29 |
ENSDART00000081465
ENSDART00000136653 ENSDART00000171232 |
ccdc106b
|
coiled-coil domain containing 106b |
| chr11_-_10850936 | 0.29 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr18_-_12416019 | 0.29 |
ENSDART00000144799
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr1_+_55140970 | 0.28 |
ENSDART00000039807
|
mb
|
myoglobin |
| chr7_+_53498152 | 0.27 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr20_-_20248408 | 0.26 |
ENSDART00000183234
|
ppp2r5ea
|
protein phosphatase 2, regulatory subunit B', epsilon isoform a |
| chr13_-_14269626 | 0.25 |
ENSDART00000079176
|
gfra4a
|
GDNF family receptor alpha 4a |
| chr9_-_40873934 | 0.24 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr14_-_4043818 | 0.24 |
ENSDART00000179870
|
snx25
|
sorting nexin 25 |
| chr19_+_37135700 | 0.21 |
ENSDART00000103159
|
smim12
|
small integral membrane protein 12 |
| chr8_+_7801060 | 0.21 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr6_-_35310224 | 0.20 |
ENSDART00000148997
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr7_+_39360797 | 0.20 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr1_+_1941031 | 0.20 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr18_+_19419120 | 0.19 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr5_-_57723929 | 0.19 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr16_-_46567136 | 0.18 |
ENSDART00000159180
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr18_+_5308392 | 0.17 |
ENSDART00000179072
|
DUT
|
deoxyuridine triphosphatase |
| chr12_-_31484677 | 0.16 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr14_-_31856819 | 0.15 |
ENSDART00000003345
|
arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr8_+_23485079 | 0.15 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr2_+_29491314 | 0.09 |
ENSDART00000181774
|
dlgap1a
|
discs, large (Drosophila) homolog-associated protein 1a |
| chr12_+_31422557 | 0.09 |
ENSDART00000153179
|
zdhhc6
|
zinc finger, DHHC-type containing 6 |
| chr1_-_45889820 | 0.09 |
ENSDART00000144735
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr22_+_21317597 | 0.08 |
ENSDART00000132605
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr11_+_36683859 | 0.08 |
ENSDART00000170102
|
si:ch211-11c3.12
|
si:ch211-11c3.12 |
| chr17_-_1407593 | 0.06 |
ENSDART00000157622
ENSDART00000159458 |
zbtb42
|
zinc finger and BTB domain containing 42 |
| chr23_-_33944597 | 0.06 |
ENSDART00000133223
|
si:dkey-190g6.2
|
si:dkey-190g6.2 |
| chr14_-_2209742 | 0.05 |
ENSDART00000054889
|
pcdh2ab5
|
protocadherin 2 alpha b 5 |
| chr2_+_25198648 | 0.05 |
ENSDART00000110922
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr22_-_36774057 | 0.04 |
ENSDART00000125048
|
acy1
|
aminoacylase 1 |
| chr3_+_12718100 | 0.04 |
ENSDART00000162343
ENSDART00000192425 |
cyp2k20
|
cytochrome P450, family 2, subfamily k, polypeptide 20 |
| chr2_-_5404466 | 0.04 |
ENSDART00000152907
|
si:ch1073-184j22.2
|
si:ch1073-184j22.2 |
| chr2_+_43469241 | 0.03 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr21_+_21679086 | 0.03 |
ENSDART00000146225
|
or125-5
|
odorant receptor, family E, subfamily 125, member 5 |
| chr18_+_7264961 | 0.03 |
ENSDART00000188461
|
CABZ01015105.1
|
|
| chr22_+_15310103 | 0.03 |
ENSDART00000145849
|
CU104797.1
|
|
| chr2_+_30465102 | 0.03 |
ENSDART00000188404
|
neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr25_-_13201458 | 0.03 |
ENSDART00000192451
|
si:ch211-194m7.4
|
si:ch211-194m7.4 |
| chr19_+_10331325 | 0.02 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr21_-_36396334 | 0.02 |
ENSDART00000183627
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr23_+_16469530 | 0.02 |
ENSDART00000132898
|
ntsr1
|
neurotensin receptor 1 (high affinity) |
| chr3_+_1096831 | 0.02 |
ENSDART00000132817
|
si:ch1073-322p19.1
|
si:ch1073-322p19.1 |
| chr15_-_37834433 | 0.02 |
ENSDART00000189748
|
si:dkey-238d18.3
|
si:dkey-238d18.3 |
| chr14_+_24042760 | 0.02 |
ENSDART00000106096
|
drd1a
|
dopamine receptor D1a |
| chr6_+_39085969 | 0.01 |
ENSDART00000004240
|
prss60.1
|
protease, serine, 60.1 |
| chr19_-_15434813 | 0.00 |
ENSDART00000019843
|
ftr55
|
finTRIM family, member 55 |
| chr9_+_11034314 | 0.00 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr18_+_32444636 | 0.00 |
ENSDART00000164223
|
v2ra1
|
vomeronasal 2 receptor, a1 |
| chr6_+_40591149 | 0.00 |
ENSDART00000189060
ENSDART00000188298 |
frs3
|
fibroblast growth factor receptor substrate 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of six4a+six4b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.3 | 1.6 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.7 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.6 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.7 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 0.7 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.7 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.1 | 0.6 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.1 | 0.5 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.3 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.1 | 0.8 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.6 | GO:0003428 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.1 | 1.0 | GO:0090279 | regulation of calcium ion import(GO:0090279) |
| 0.1 | 0.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 2.3 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.8 | GO:0035459 | cargo loading into vesicle(GO:0035459) |
| 0.0 | 0.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.6 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0060004 | reflex(GO:0060004) vestibular reflex(GO:0060005) |
| 0.0 | 1.1 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.6 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.5 | GO:1902285 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.3 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.6 | GO:0097192 | signal transduction in absence of ligand(GO:0038034) extrinsic apoptotic signaling pathway in absence of ligand(GO:0097192) |
| 0.0 | 0.5 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 1.3 | GO:0071560 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.2 | 0.7 | GO:0000941 | condensed nuclear chromosome inner kinetochore(GO:0000941) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.4 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.3 | GO:1990513 | CLOCK-BMAL transcription complex(GO:1990513) |
| 0.0 | 0.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.0 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.1 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0003913 | DNA photolyase activity(GO:0003913) |
| 0.2 | 0.5 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.1 | 1.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.7 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 1.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.1 | 0.8 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.8 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 6.9 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.6 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.3 | GO:0019825 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.3 | GO:0016411 | acylglycerol O-acyltransferase activity(GO:0016411) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 1.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.4 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.4 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |