Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
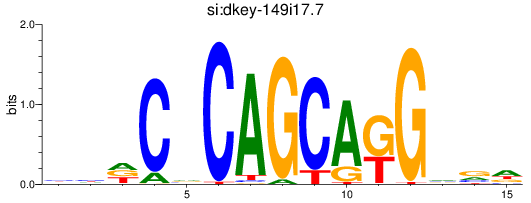
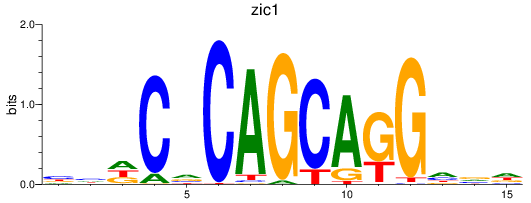
Results for si:dkey-149i17.7_zic1
Z-value: 0.82
Transcription factors associated with si:dkey-149i17.7_zic1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-149i17.7
|
ENSDARG00000094684 | si_dkey-149i17.7 |
|
zic1
|
ENSDARG00000015567 | zic family member 1 (odd-paired homolog, Drosophila) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-149i17.7 | dr11_v1_chr2_+_24885987_24885987 | -0.19 | 4.5e-01 | Click! |
| zic1 | dr11_v1_chr24_-_4973765_4973765 | 0.06 | 8.2e-01 | Click! |
Activity profile of si:dkey-149i17.7_zic1 motif
Sorted Z-values of si:dkey-149i17.7_zic1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_6250317 | 3.26 |
ENSDART00000180416
|
tuba4l
|
tubulin, alpha 4 like |
| chr8_+_14987006 | 3.13 |
ENSDART00000045038
|
fnbp1l
|
formin binding protein 1-like |
| chr2_+_41526904 | 2.88 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr20_+_20731633 | 2.26 |
ENSDART00000191952
ENSDART00000165224 |
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr6_+_10338554 | 2.19 |
ENSDART00000186936
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr15_+_20239141 | 2.12 |
ENSDART00000101152
ENSDART00000152473 |
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr22_-_20924564 | 2.05 |
ENSDART00000100642
ENSDART00000032770 |
ell
|
elongation factor RNA polymerase II |
| chr15_+_45591669 | 2.03 |
ENSDART00000157459
|
atg16l1
|
ATG16 autophagy related 16-like 1 (S. cerevisiae) |
| chr16_+_31511739 | 1.94 |
ENSDART00000049420
|
ndrg1b
|
N-myc downstream regulated 1b |
| chr24_-_34680956 | 1.92 |
ENSDART00000171009
|
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr21_-_22357545 | 1.89 |
ENSDART00000134320
|
skp2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr11_+_14333441 | 1.88 |
ENSDART00000171969
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr17_+_24064014 | 1.87 |
ENSDART00000182782
ENSDART00000139063 ENSDART00000132755 |
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr22_-_14247276 | 1.76 |
ENSDART00000033332
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr18_+_3243292 | 1.75 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr12_-_1034383 | 1.74 |
ENSDART00000152455
ENSDART00000152346 |
polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr14_-_34633960 | 1.72 |
ENSDART00000128869
ENSDART00000179977 |
afap1l1a
|
actin filament associated protein 1-like 1a |
| chr14_+_35428152 | 1.70 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr23_+_40139765 | 1.67 |
ENSDART00000185376
|
gpsm2l
|
G protein signaling modulator 2, like |
| chr4_-_4535189 | 1.67 |
ENSDART00000057519
|
zgc:194209
|
zgc:194209 |
| chr24_+_25822999 | 1.63 |
ENSDART00000109809
|
sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr6_+_29923593 | 1.60 |
ENSDART00000169687
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr18_+_3169579 | 1.60 |
ENSDART00000164724
ENSDART00000186340 ENSDART00000181247 ENSDART00000168056 |
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr10_+_37182626 | 1.56 |
ENSDART00000137636
|
ksr1a
|
kinase suppressor of ras 1a |
| chr19_-_10915898 | 1.54 |
ENSDART00000163179
|
pip5k1aa
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, a |
| chr3_+_54047342 | 1.50 |
ENSDART00000178486
|
olfm2a
|
olfactomedin 2a |
| chr18_+_39487486 | 1.48 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr11_-_44999858 | 1.48 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr12_-_35393211 | 1.38 |
ENSDART00000137139
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr5_-_32332560 | 1.31 |
ENSDART00000083862
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr14_+_30285613 | 1.28 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr9_+_33340311 | 1.26 |
ENSDART00000140064
|
ddx3a
|
DEAD (Asp-Glu-Ala-Asp) box helicase 3a |
| chr7_-_59514547 | 1.25 |
ENSDART00000168457
|
slx1b
|
SLX1 homolog B, structure-specific endonuclease subunit |
| chr7_+_39418869 | 1.25 |
ENSDART00000169195
|
CT030188.1
|
|
| chr24_+_14801844 | 1.24 |
ENSDART00000141620
|
pi15a
|
peptidase inhibitor 15a |
| chr4_-_2162688 | 1.23 |
ENSDART00000148900
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr11_-_669558 | 1.23 |
ENSDART00000173450
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr21_-_42831033 | 1.21 |
ENSDART00000160998
|
stk10
|
serine/threonine kinase 10 |
| chr19_-_1002959 | 1.20 |
ENSDART00000168138
|
ehmt2
|
euchromatic histone-lysine N-methyltransferase 2 |
| chr23_-_32334208 | 1.19 |
ENSDART00000053472
|
rnf41
|
ring finger protein 41 |
| chr6_+_3373665 | 1.19 |
ENSDART00000134133
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr25_+_3759553 | 1.16 |
ENSDART00000180601
ENSDART00000055845 ENSDART00000157050 ENSDART00000153905 |
thoc5
|
THO complex 5 |
| chr12_-_33789006 | 1.13 |
ENSDART00000034550
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr9_-_28255029 | 1.13 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr2_+_44977889 | 1.12 |
ENSDART00000144024
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr11_-_669270 | 1.11 |
ENSDART00000172834
|
pparg
|
peroxisome proliferator-activated receptor gamma |
| chr18_+_907266 | 1.09 |
ENSDART00000171729
|
pkma
|
pyruvate kinase M1/2a |
| chr16_+_50755133 | 1.07 |
ENSDART00000029283
|
IGLON5
|
zgc:110372 |
| chr17_-_25861787 | 1.07 |
ENSDART00000182503
|
BX000364.5
|
|
| chr3_-_2613990 | 1.06 |
ENSDART00000137102
|
si:dkey-217f16.6
|
si:dkey-217f16.6 |
| chr2_-_47957673 | 1.06 |
ENSDART00000056305
|
fzd8b
|
frizzled class receptor 8b |
| chr5_-_20921677 | 1.06 |
ENSDART00000158030
|
si:ch211-225b11.4
|
si:ch211-225b11.4 |
| chr12_-_33789218 | 1.04 |
ENSDART00000193258
|
llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr16_-_25680666 | 1.04 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr18_+_34181655 | 1.04 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr19_-_29294457 | 1.03 |
ENSDART00000130815
ENSDART00000103437 |
e2f3
|
E2F transcription factor 3 |
| chr11_+_24034345 | 1.01 |
ENSDART00000128309
|
zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr12_-_13729263 | 0.98 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr17_+_13664442 | 0.98 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr7_-_18881358 | 0.98 |
ENSDART00000021502
|
mllt3
|
MLLT3, super elongation complex subunit |
| chr24_-_26854032 | 0.97 |
ENSDART00000087991
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr21_+_76739 | 0.96 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr16_+_50741154 | 0.96 |
ENSDART00000101627
|
IGLON5
|
zgc:110372 |
| chr6_-_33707278 | 0.94 |
ENSDART00000188103
|
mast2
|
microtubule associated serine/threonine kinase 2 |
| chr6_+_19933763 | 0.94 |
ENSDART00000166192
|
pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr16_+_25296389 | 0.94 |
ENSDART00000114528
|
tbc1d31
|
TBC1 domain family, member 31 |
| chr6_-_3978919 | 0.92 |
ENSDART00000167753
|
slc25a12
|
solute carrier family 25 (aspartate/glutamate carrier), member 12 |
| chr10_-_2526526 | 0.92 |
ENSDART00000191726
ENSDART00000192767 |
CU856539.1
|
|
| chr17_-_52941606 | 0.91 |
ENSDART00000155310
|
zgc:154061
|
zgc:154061 |
| chr5_-_66823750 | 0.90 |
ENSDART00000041441
ENSDART00000112488 |
stip1
|
stress-induced phosphoprotein 1 |
| chr17_+_24446353 | 0.90 |
ENSDART00000140467
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr21_-_4793686 | 0.90 |
ENSDART00000158232
|
notch1a
|
notch 1a |
| chr23_-_24226533 | 0.87 |
ENSDART00000109134
|
plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr18_+_35229115 | 0.83 |
ENSDART00000129624
ENSDART00000184596 |
tbrg1
|
transforming growth factor beta regulator 1 |
| chr17_+_24446705 | 0.83 |
ENSDART00000163221
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr24_-_41267184 | 0.83 |
ENSDART00000063504
|
xylb
|
xylulokinase homolog (H. influenzae) |
| chr17_+_24446533 | 0.82 |
ENSDART00000131417
|
ugp2b
|
UDP-glucose pyrophosphorylase 2b |
| chr2_-_15041846 | 0.81 |
ENSDART00000139050
|
si:dkey-10f21.4
|
si:dkey-10f21.4 |
| chr8_+_47219107 | 0.81 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr18_-_22094102 | 0.81 |
ENSDART00000100904
|
pard6a
|
par-6 family cell polarity regulator alpha |
| chr9_+_29430432 | 0.78 |
ENSDART00000125632
|
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr18_+_3634652 | 0.76 |
ENSDART00000159913
|
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr20_-_18794789 | 0.73 |
ENSDART00000003834
|
ccm2
|
cerebral cavernous malformation 2 |
| chr24_-_17400143 | 0.73 |
ENSDART00000134947
|
cul1b
|
cullin 1b |
| chr4_-_11024767 | 0.72 |
ENSDART00000067261
ENSDART00000167631 |
tmtc2a
|
transmembrane and tetratricopeptide repeat containing 2a |
| chr9_-_21238159 | 0.71 |
ENSDART00000146764
ENSDART00000102143 |
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr13_+_9559461 | 0.71 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr2_-_37134169 | 0.71 |
ENSDART00000146123
ENSDART00000146533 ENSDART00000040427 |
elavl1a
|
ELAV like RNA binding protein 1a |
| chr11_-_10850936 | 0.70 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr1_+_494297 | 0.69 |
ENSDART00000108579
ENSDART00000146732 |
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr8_+_30117297 | 0.69 |
ENSDART00000077554
|
fancc
|
Fanconi anemia, complementation group C |
| chr2_+_11031360 | 0.68 |
ENSDART00000180020
ENSDART00000145093 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr24_-_17400472 | 0.67 |
ENSDART00000024691
|
cul1b
|
cullin 1b |
| chr23_-_40194732 | 0.67 |
ENSDART00000164931
|
tgm1l2
|
transglutaminase 1 like 2 |
| chr11_-_36230146 | 0.67 |
ENSDART00000135888
ENSDART00000189782 |
rrp9
|
ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) |
| chr5_+_37744625 | 0.67 |
ENSDART00000014031
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr23_+_28718863 | 0.66 |
ENSDART00000078156
|
srm
|
spermidine synthase |
| chr11_-_28747 | 0.66 |
ENSDART00000172956
|
sp1
|
sp1 transcription factor |
| chr9_+_8364553 | 0.64 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr11_+_41838801 | 0.64 |
ENSDART00000014871
|
akr7a3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr13_+_40334727 | 0.64 |
ENSDART00000074950
|
slc25a28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr21_+_33311622 | 0.63 |
ENSDART00000163808
|
si:ch211-151g22.1
|
si:ch211-151g22.1 |
| chr3_-_31254979 | 0.63 |
ENSDART00000130280
|
apnl
|
actinoporin-like protein |
| chr21_+_26991198 | 0.62 |
ENSDART00000065397
|
fkbp2
|
FK506 binding protein 2 |
| chr18_+_1615 | 0.62 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr23_+_1349277 | 0.59 |
ENSDART00000173133
ENSDART00000179877 |
utrn
|
utrophin |
| chr22_-_37738203 | 0.58 |
ENSDART00000143190
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr5_-_72178739 | 0.55 |
ENSDART00000050971
|
rab14l
|
RAB14, member RAS oncogene family, like |
| chr15_+_29728377 | 0.52 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr2_+_25378457 | 0.51 |
ENSDART00000089108
|
fndc3ba
|
fibronectin type III domain containing 3Ba |
| chr21_-_40557281 | 0.51 |
ENSDART00000172327
|
taok1b
|
TAO kinase 1b |
| chr23_-_4704938 | 0.50 |
ENSDART00000067293
|
cnbpa
|
CCHC-type zinc finger, nucleic acid binding protein a |
| chr14_+_23709543 | 0.50 |
ENSDART00000136909
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr20_-_18731268 | 0.50 |
ENSDART00000183893
|
enpp5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 |
| chr8_+_26874924 | 0.50 |
ENSDART00000141794
|
rimkla
|
ribosomal modification protein rimK-like family member A |
| chr13_+_31172833 | 0.50 |
ENSDART00000176378
|
CR931802.3
|
|
| chr7_-_7692723 | 0.49 |
ENSDART00000183352
|
aadat
|
aminoadipate aminotransferase |
| chr8_-_20838342 | 0.49 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr16_-_40426837 | 0.49 |
ENSDART00000193690
|
plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr8_+_22277198 | 0.48 |
ENSDART00000005989
|
dffb
|
DNA fragmentation factor, beta polypeptide (caspase-activated DNase) |
| chr8_-_25600888 | 0.48 |
ENSDART00000193860
|
stk38a
|
serine/threonine kinase 38a |
| chr21_+_3901775 | 0.48 |
ENSDART00000053609
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr13_-_41482064 | 0.47 |
ENSDART00000188322
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr21_-_13230925 | 0.47 |
ENSDART00000023834
|
setb
|
SET nuclear proto-oncogene b |
| chr17_+_33296852 | 0.47 |
ENSDART00000154580
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_-_23848012 | 0.47 |
ENSDART00000146104
ENSDART00000175108 |
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr17_-_15667198 | 0.46 |
ENSDART00000142972
ENSDART00000132571 ENSDART00000189936 |
manea
|
mannosidase, endo-alpha |
| chr14_+_35405518 | 0.45 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| chr14_-_47882706 | 0.45 |
ENSDART00000188772
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr7_-_7692992 | 0.45 |
ENSDART00000192619
|
aadat
|
aminoadipate aminotransferase |
| chr4_+_6869847 | 0.45 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr19_-_48010490 | 0.44 |
ENSDART00000159938
|
FBXL19
|
zgc:158376 |
| chr19_-_17996162 | 0.44 |
ENSDART00000150928
ENSDART00000104491 |
ints8
|
integrator complex subunit 8 |
| chr3_-_21061931 | 0.43 |
ENSDART00000036741
|
fam57ba
|
family with sequence similarity 57, member Ba |
| chr3_+_46479913 | 0.43 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr13_-_25719628 | 0.43 |
ENSDART00000135383
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr6_+_16493755 | 0.42 |
ENSDART00000126318
|
si:ch73-193c12.2
|
si:ch73-193c12.2 |
| chr9_+_34380299 | 0.42 |
ENSDART00000131705
|
lamp1
|
lysosomal-associated membrane protein 1 |
| chr15_+_47083163 | 0.42 |
ENSDART00000097465
|
cep57
|
centrosomal protein 57 |
| chr5_-_5326010 | 0.41 |
ENSDART00000161946
|
pbx3a
|
pre-B-cell leukemia homeobox 3a |
| chr25_+_37435720 | 0.41 |
ENSDART00000164390
|
chmp1a
|
charged multivesicular body protein 1A |
| chr5_-_45651548 | 0.41 |
ENSDART00000097645
|
NPFFR2
|
neuropeptide FF receptor 2a |
| chr19_-_17996336 | 0.40 |
ENSDART00000186143
ENSDART00000080751 |
ints8
|
integrator complex subunit 8 |
| chr8_+_54055390 | 0.40 |
ENSDART00000102696
|
magi1a
|
membrane associated guanylate kinase, WW and PDZ domain containing 1a |
| chr24_+_36018164 | 0.39 |
ENSDART00000182815
ENSDART00000126941 |
gnal2
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type 2 |
| chr24_+_39227519 | 0.39 |
ENSDART00000184611
ENSDART00000193494 ENSDART00000190728 ENSDART00000168705 |
MPRIP
|
si:ch73-103b11.2 |
| chr21_-_13231101 | 0.38 |
ENSDART00000190943
|
setb
|
SET nuclear proto-oncogene b |
| chr5_-_32074534 | 0.38 |
ENSDART00000112342
ENSDART00000135855 |
arpc5lb
|
actin related protein 2/3 complex, subunit 5-like, b |
| chr15_-_25613114 | 0.38 |
ENSDART00000182431
ENSDART00000187405 |
taok1a
|
TAO kinase 1a |
| chr14_-_4043818 | 0.38 |
ENSDART00000179870
|
snx25
|
sorting nexin 25 |
| chr8_-_4574328 | 0.37 |
ENSDART00000090731
|
dhx37
|
DEAH (Asp-Glu-Ala-His) box polypeptide 37 |
| chr14_-_30876708 | 0.37 |
ENSDART00000147597
|
ubl3b
|
ubiquitin-like 3b |
| chr21_+_3093419 | 0.37 |
ENSDART00000162520
|
SHC3
|
SHC adaptor protein 3 |
| chr17_-_27048537 | 0.36 |
ENSDART00000050018
ENSDART00000193861 |
cnksr1
|
connector enhancer of kinase suppressor of Ras 1 |
| chr10_+_589501 | 0.36 |
ENSDART00000188415
|
LO018557.1
|
|
| chr23_+_27789795 | 0.36 |
ENSDART00000141458
|
kmt2d
|
lysine (K)-specific methyltransferase 2D |
| chr23_-_25126003 | 0.36 |
ENSDART00000034953
|
idh3g
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr7_-_5396154 | 0.36 |
ENSDART00000172980
|
arhgef11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr13_-_44843819 | 0.36 |
ENSDART00000140442
|
si:dkeyp-2e4.3
|
si:dkeyp-2e4.3 |
| chr13_-_24396199 | 0.35 |
ENSDART00000181093
|
tbp
|
TATA box binding protein |
| chr25_-_27729046 | 0.35 |
ENSDART00000131437
|
zgc:153935
|
zgc:153935 |
| chr2_+_51028269 | 0.34 |
ENSDART00000161254
|
eef1da
|
eukaryotic translation elongation factor 1 delta a (guanine nucleotide exchange protein) |
| chr16_+_33142734 | 0.34 |
ENSDART00000138244
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr16_+_2905150 | 0.34 |
ENSDART00000109980
|
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr5_-_21044693 | 0.32 |
ENSDART00000140298
|
si:dkey-13n15.2
|
si:dkey-13n15.2 |
| chr3_+_52078798 | 0.32 |
ENSDART00000156882
|
si:dkey-88e12.3
|
si:dkey-88e12.3 |
| chr3_-_40162843 | 0.32 |
ENSDART00000129664
ENSDART00000025285 |
drg2
|
developmentally regulated GTP binding protein 2 |
| chr25_-_893464 | 0.31 |
ENSDART00000159321
|
znf609a
|
zinc finger protein 609a |
| chr13_+_10799380 | 0.31 |
ENSDART00000126427
|
lrpprc
|
leucine-rich pentatricopeptide repeat containing |
| chr25_-_16782394 | 0.30 |
ENSDART00000019413
|
ndufa9a
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9a |
| chr5_-_37044262 | 0.30 |
ENSDART00000166670
|
si:dkeyp-110c7.8
|
si:dkeyp-110c7.8 |
| chr19_-_27391179 | 0.30 |
ENSDART00000181108
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr20_-_54522660 | 0.30 |
ENSDART00000059872
|
ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr22_+_35275206 | 0.30 |
ENSDART00000112234
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr7_-_40959867 | 0.29 |
ENSDART00000174009
|
rbm33a
|
RNA binding motif protein 33a |
| chr3_+_27722355 | 0.29 |
ENSDART00000132761
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr1_+_45085194 | 0.29 |
ENSDART00000193863
|
si:ch211-151p13.8
|
si:ch211-151p13.8 |
| chr24_+_31379318 | 0.28 |
ENSDART00000183707
|
cpne3
|
copine III |
| chr22_+_30038459 | 0.28 |
ENSDART00000177014
|
AL845330.1
|
|
| chr12_+_4225199 | 0.28 |
ENSDART00000042277
|
mapk7
|
mitogen-activated protein kinase 7 |
| chr17_-_3303805 | 0.28 |
ENSDART00000169136
|
CABZ01007222.1
|
|
| chr19_+_1999838 | 0.28 |
ENSDART00000166669
|
btd
|
biotinidase |
| chr19_-_32641725 | 0.26 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr17_+_23926796 | 0.26 |
ENSDART00000021177
|
pex13
|
peroxisomal biogenesis factor 13 |
| chr7_+_67429185 | 0.26 |
ENSDART00000162553
ENSDART00000178646 |
kars
|
lysyl-tRNA synthetase |
| chr8_-_11770092 | 0.26 |
ENSDART00000091684
|
anapc7
|
anaphase promoting complex subunit 7 |
| chr13_-_33198150 | 0.25 |
ENSDART00000137576
|
trip11
|
thyroid hormone receptor interactor 11 |
| chr1_-_59313465 | 0.25 |
ENSDART00000158067
ENSDART00000159419 |
txndc11
|
thioredoxin domain containing 11 |
| chr18_-_45736 | 0.25 |
ENSDART00000148373
ENSDART00000148950 |
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr22_+_2648931 | 0.25 |
ENSDART00000106357
|
zgc:110821
|
zgc:110821 |
| chr10_+_35329751 | 0.24 |
ENSDART00000148043
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr1_-_20068155 | 0.24 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr11_-_28224 | 0.24 |
ENSDART00000124104
|
sp1
|
sp1 transcription factor |
| chr10_-_18492617 | 0.24 |
ENSDART00000179936
ENSDART00000193799 ENSDART00000193198 |
si:dkey-28o19.1
|
si:dkey-28o19.1 |
| chr8_-_14126646 | 0.24 |
ENSDART00000027225
|
bgna
|
biglycan a |
| chr8_+_31777633 | 0.23 |
ENSDART00000142519
|
oxct1a
|
3-oxoacid CoA transferase 1a |
| chr2_-_39558643 | 0.23 |
ENSDART00000139860
ENSDART00000145231 ENSDART00000141721 |
cbln7
|
cerebellin 7 |
| chr16_+_26706519 | 0.22 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr10_-_1523253 | 0.22 |
ENSDART00000179510
ENSDART00000176548 ENSDART00000180368 ENSDART00000185270 |
WDR70
|
WD repeat domain 70 |
| chr25_+_16080181 | 0.21 |
ENSDART00000061753
|
far1
|
fatty acyl CoA reductase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-149i17.7_zic1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.6 | 3.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.5 | 2.5 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.4 | 2.2 | GO:0010719 | photoreceptor cell morphogenesis(GO:0008594) negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.4 | 2.0 | GO:0039694 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.3 | 2.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 1.0 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.3 | 0.9 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.3 | 1.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.3 | 1.9 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 0.8 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 1.1 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 1.2 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.2 | 0.9 | GO:0032655 | interleukin-12 production(GO:0032615) regulation of interleukin-12 production(GO:0032655) |
| 0.2 | 0.5 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.2 | 0.5 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.2 | 2.3 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.2 | 1.3 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.4 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.8 | GO:0044364 | killing of cells of other organism(GO:0031640) disruption of cells of other organism(GO:0044364) disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.1 | 0.5 | GO:0090387 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.1 | 1.9 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.5 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.2 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 1.0 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.1 | 0.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 1.6 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.3 | GO:1901004 | ubiquinone-6 metabolic process(GO:1901004) ubiquinone-6 biosynthetic process(GO:1901006) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.9 | GO:0015813 | acidic amino acid transport(GO:0015800) aspartate transport(GO:0015810) L-glutamate transport(GO:0015813) malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.1 | GO:1903729 | regulation of plasma membrane organization(GO:1903729) |
| 0.1 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.6 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 0.7 | GO:0008216 | spermidine metabolic process(GO:0008216) |
| 0.1 | 1.6 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.0 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.5 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 1.1 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 1.0 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.1 | 1.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 1.0 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.7 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.2 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.1 | 1.4 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.0 | 0.8 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.2 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.4 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:1900182 | positive regulation of protein localization to nucleus(GO:1900182) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.4 | GO:0003422 | growth plate cartilage morphogenesis(GO:0003422) chondrocyte intercalation involved in growth plate cartilage morphogenesis(GO:0003428) |
| 0.0 | 0.7 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0061033 | lung growth(GO:0060437) secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.0 | 1.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.0 | 0.8 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.7 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 1.1 | GO:0072114 | pronephros morphogenesis(GO:0072114) |
| 0.0 | 0.1 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.1 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.3 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.7 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 1.9 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.8 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) quinolinate metabolic process(GO:0046874) |
| 0.0 | 0.9 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.0 | 1.5 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 1.1 | GO:0006096 | glycolytic process(GO:0006096) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.2 | GO:0019730 | antimicrobial humoral response(GO:0019730) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.3 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.1 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.0 | 1.3 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.0 | 1.7 | GO:0045786 | negative regulation of cell cycle(GO:0045786) |
| 0.0 | 3.1 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.3 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 0.7 | GO:0055088 | lipid homeostasis(GO:0055088) |
| 0.0 | 0.7 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.4 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.3 | 0.8 | GO:0044218 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.3 | 2.2 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.2 | 1.3 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 0.1 | 1.7 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.9 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.0 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.7 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.1 | 0.9 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.2 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.7 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.7 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.2 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.6 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.1 | 0.2 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 2.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 2.2 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.8 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 3.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 2.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.7 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 3.3 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0003983 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.5 | 1.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.5 | 2.3 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.4 | 1.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.3 | 1.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.3 | 1.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.2 | 2.9 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.2 | 0.5 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 1.2 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.2 | 0.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 0.5 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 0.5 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) citrate-L-glutamate ligase activity(GO:0072591) |
| 0.2 | 0.8 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.2 | 2.8 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 1.7 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.1 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.9 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 1.9 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 2.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.1 | 2.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.2 | GO:0001734 | mRNA (N6-adenosine)-methyltransferase activity(GO:0001734) |
| 0.1 | 0.9 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0043734 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 1.0 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 1.0 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.9 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.1 | 1.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.7 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.0 | 0.2 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 2.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.0 | 0.9 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 1.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.8 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 1.9 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.4 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.2 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 1.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 3.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 0.5 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.4 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.6 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.9 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.7 | GO:0004536 | deoxyribonuclease activity(GO:0004536) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 1.9 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.2 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 2.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.6 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.3 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 1.0 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.2 | 1.3 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.2 | 0.8 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.7 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.5 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 2.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 1.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.0 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 2.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 3.0 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.1 | 0.8 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.2 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 2.3 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.6 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION BY GLUCAGON LIKE PEPTIDE1 | Genes involved in Regulation of Insulin Secretion by Glucagon-like Peptide-1 |
| 0.0 | 0.7 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.9 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.3 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |