Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
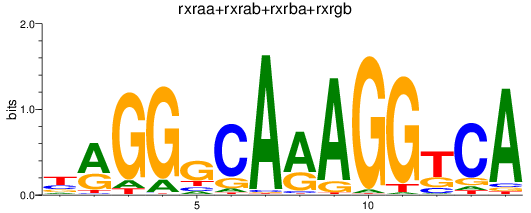
Results for rxraa+rxrab+rxrba+rxrgb
Z-value: 0.99
Transcription factors associated with rxraa+rxrab+rxrba+rxrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rxrgb
|
ENSDARG00000004697 | retinoid X receptor, gamma b |
|
rxrab
|
ENSDARG00000035127 | retinoid x receptor, alpha b |
|
rxraa
|
ENSDARG00000057737 | retinoid X receptor, alpha a |
|
rxrba
|
ENSDARG00000078954 | retinoid x receptor, beta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rxrgb | dr11_v1_chr20_+_33875256_33875256 | -0.68 | 1.8e-03 | Click! |
| rxrba | dr11_v1_chr19_-_7272921_7273057 | 0.38 | 1.2e-01 | Click! |
| rxrab | dr11_v1_chr5_-_64511428_64511428 | 0.20 | 4.3e-01 | Click! |
| rxraa | dr11_v1_chr21_+_17880511_17880511 | -0.10 | 7.1e-01 | Click! |
Activity profile of rxraa+rxrab+rxrba+rxrgb motif
Sorted Z-values of rxraa+rxrab+rxrba+rxrgb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_154556 | 3.93 |
ENSDART00000193153
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr11_+_18175893 | 3.41 |
ENSDART00000177625
|
zgc:173545
|
zgc:173545 |
| chr11_+_18037729 | 3.35 |
ENSDART00000111624
|
zgc:175135
|
zgc:175135 |
| chr11_+_18130300 | 3.31 |
ENSDART00000169146
|
zgc:175135
|
zgc:175135 |
| chr11_+_18157260 | 3.27 |
ENSDART00000144659
|
zgc:173545
|
zgc:173545 |
| chr11_+_18053333 | 3.27 |
ENSDART00000075750
|
zgc:175135
|
zgc:175135 |
| chr20_+_54304800 | 2.90 |
ENSDART00000121661
|
zp2.6
|
zona pellucida glycoprotein 2, tandem duplicate 6 |
| chr20_+_54295213 | 2.52 |
ENSDART00000074085
|
zp2.3
|
zona pellucida glycoprotein 2, tandem duplicate 3 |
| chr20_+_54312970 | 2.51 |
ENSDART00000024598
ENSDART00000193172 |
zp2.5
|
zona pellucida glycoprotein 2, tandem duplicate 5 |
| chr20_+_54309148 | 2.48 |
ENSDART00000099360
|
zp2.1
|
zona pellucida glycoprotein 2, tandem duplicate 1 |
| chr20_+_54299419 | 2.47 |
ENSDART00000056089
ENSDART00000193107 |
si:zfos-1505d6.3
|
si:zfos-1505d6.3 |
| chr19_-_27564458 | 2.41 |
ENSDART00000123155
|
si:dkeyp-46h3.6
|
si:dkeyp-46h3.6 |
| chr21_+_25777425 | 2.34 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr6_+_153146 | 2.33 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr9_+_33216945 | 2.31 |
ENSDART00000134029
|
si:ch211-125e6.12
|
si:ch211-125e6.12 |
| chr7_-_24520866 | 2.17 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr9_+_8380728 | 2.14 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr19_-_27564980 | 2.13 |
ENSDART00000171967
|
si:dkeyp-46h3.8
|
si:dkeyp-46h3.8 |
| chr23_+_2740741 | 1.99 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr22_-_7050 | 1.68 |
ENSDART00000127829
|
atad3
|
ATPase family, AAA domain containing 3 |
| chr15_-_28908027 | 1.64 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr3_+_27770110 | 1.64 |
ENSDART00000017962
|
eci1
|
enoyl-CoA delta isomerase 1 |
| chr15_-_28907709 | 1.58 |
ENSDART00000017268
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr22_-_24992532 | 1.52 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr20_-_14114078 | 1.39 |
ENSDART00000168434
ENSDART00000104032 |
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr8_-_2529878 | 1.38 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr24_-_5691956 | 1.36 |
ENSDART00000189112
|
dia1b
|
deleted in autism 1b |
| chr22_-_5171362 | 1.30 |
ENSDART00000124889
|
tnfaip8l1
|
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr23_+_30730121 | 1.26 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr16_+_27614989 | 1.25 |
ENSDART00000005625
|
glipr2l
|
GLI pathogenesis-related 2, like |
| chr3_-_36419641 | 1.21 |
ENSDART00000173545
|
cog1
|
component of oligomeric golgi complex 1 |
| chr10_+_24692076 | 1.20 |
ENSDART00000181600
|
tpte
|
transmembrane phosphatase with tensin homology |
| chr8_+_2530065 | 1.17 |
ENSDART00000063943
|
mrpl40
|
mitochondrial ribosomal protein L40 |
| chr2_-_26476030 | 1.17 |
ENSDART00000145262
ENSDART00000132125 |
acadm
|
acyl-CoA dehydrogenase medium chain |
| chr14_+_34495216 | 1.16 |
ENSDART00000147756
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr10_+_15340768 | 1.14 |
ENSDART00000046274
ENSDART00000168909 |
trappc13
|
trafficking protein particle complex 13 |
| chr18_-_158541 | 1.08 |
ENSDART00000188914
ENSDART00000191052 |
trpm7
|
transient receptor potential cation channel, subfamily M, member 7 |
| chr3_-_34561624 | 1.04 |
ENSDART00000129313
|
sept9a
|
septin 9a |
| chr16_-_39131666 | 1.01 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr18_+_1615 | 1.01 |
ENSDART00000082450
|
homer2
|
homer scaffolding protein 2 |
| chr23_+_30736895 | 1.01 |
ENSDART00000042944
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr15_-_6946286 | 1.01 |
ENSDART00000019330
|
ech1
|
enoyl CoA hydratase 1, peroxisomal |
| chr9_+_22782027 | 1.01 |
ENSDART00000090816
|
rif1
|
replication timing regulatory factor 1 |
| chr7_+_24520518 | 0.99 |
ENSDART00000173604
|
btr09
|
bloodthirsty-related gene family, member 9 |
| chr24_-_32522587 | 0.97 |
ENSDART00000048968
ENSDART00000143781 |
EIF1B
|
zgc:56676 |
| chr3_+_7808459 | 0.97 |
ENSDART00000162374
|
hook2
|
hook microtubule-tethering protein 2 |
| chr16_-_19568388 | 0.95 |
ENSDART00000141616
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr11_+_45287541 | 0.94 |
ENSDART00000165321
ENSDART00000173116 |
pycr1b
|
pyrroline-5-carboxylate reductase 1b |
| chr18_-_34549721 | 0.91 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr24_-_26820698 | 0.89 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr6_+_40952031 | 0.89 |
ENSDART00000189219
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr8_-_16464453 | 0.88 |
ENSDART00000098691
|
rnf11b
|
ring finger protein 11b |
| chr9_-_25255490 | 0.88 |
ENSDART00000141502
|
htr2aa
|
5-hydroxytryptamine (serotonin) receptor 2A, genome duplicate a |
| chr25_-_35113891 | 0.87 |
ENSDART00000190724
|
zgc:165555
|
zgc:165555 |
| chr23_-_27050083 | 0.86 |
ENSDART00000142324
ENSDART00000133249 ENSDART00000138751 ENSDART00000128718 |
zgc:66440
|
zgc:66440 |
| chr18_+_27515640 | 0.82 |
ENSDART00000181593
|
tp53i11b
|
tumor protein p53 inducible protein 11b |
| chr22_-_26834043 | 0.80 |
ENSDART00000087202
|
si:dkey-44g23.5
|
si:dkey-44g23.5 |
| chr23_+_2914577 | 0.80 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr19_-_3056235 | 0.80 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr14_+_1719367 | 0.79 |
ENSDART00000157696
|
trpc7b
|
transient receptor potential cation channel, subfamily C, member 7b |
| chr14_-_9085349 | 0.79 |
ENSDART00000054710
|
polr1d
|
polymerase (RNA) I polypeptide D |
| chr12_-_1405228 | 0.78 |
ENSDART00000152127
|
MED9
|
si:ch73-105m5.1 |
| chr20_-_42241150 | 0.77 |
ENSDART00000142791
|
nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr8_+_11687254 | 0.76 |
ENSDART00000042040
|
atp2a2a
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 2a |
| chr3_+_34149337 | 0.75 |
ENSDART00000006091
|
carm1
|
coactivator-associated arginine methyltransferase 1 |
| chr14_+_989733 | 0.75 |
ENSDART00000161487
ENSDART00000127317 |
si:ch73-308l14.2
|
si:ch73-308l14.2 |
| chr14_+_30340251 | 0.74 |
ENSDART00000148448
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr10_+_35417099 | 0.74 |
ENSDART00000063398
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr10_+_11265387 | 0.72 |
ENSDART00000038888
|
hsdl2
|
hydroxysteroid dehydrogenase like 2 |
| chr3_-_36440705 | 0.72 |
ENSDART00000162875
|
rogdi
|
rogdi homolog (Drosophila) |
| chr16_-_19568795 | 0.71 |
ENSDART00000185141
|
abcb5
|
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr19_+_46113828 | 0.71 |
ENSDART00000159331
ENSDART00000161826 |
rbm24a
|
RNA binding motif protein 24a |
| chr13_-_25199260 | 0.71 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr6_-_49547680 | 0.71 |
ENSDART00000169678
|
ppp4r1l
|
protein phosphatase 4, regulatory subunit 1-like |
| chr18_+_44769027 | 0.69 |
ENSDART00000145190
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr22_-_10774735 | 0.69 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr18_+_30441740 | 0.68 |
ENSDART00000189074
|
gse1
|
Gse1 coiled-coil protein |
| chr20_-_32981053 | 0.67 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr18_+_39487486 | 0.67 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase long chain |
| chr25_-_25575717 | 0.66 |
ENSDART00000067138
|
hic1l
|
hypermethylated in cancer 1 like |
| chr13_-_8692860 | 0.65 |
ENSDART00000058107
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr11_+_24313931 | 0.65 |
ENSDART00000017599
ENSDART00000166045 |
rem1
|
RAS (RAD and GEM)-like GTP-binding 1 |
| chr24_+_39137001 | 0.65 |
ENSDART00000181086
ENSDART00000183724 ENSDART00000193466 |
tbc1d24
|
TBC1 domain family, member 24 |
| chr9_+_50000504 | 0.63 |
ENSDART00000164409
ENSDART00000165451 ENSDART00000166509 |
slc38a11
|
solute carrier family 38, member 11 |
| chr1_-_45213565 | 0.63 |
ENSDART00000145757
|
ddx39aa
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39Aa |
| chr2_-_57076687 | 0.63 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr24_-_21172122 | 0.63 |
ENSDART00000154259
|
atp6v1ab
|
ATPase H+ transporting V1 subunit Ab |
| chr20_-_1635922 | 0.62 |
ENSDART00000181502
|
CR846082.1
|
|
| chr7_+_22718251 | 0.62 |
ENSDART00000027718
ENSDART00000143341 |
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr21_-_31238244 | 0.61 |
ENSDART00000159678
|
tpst1l
|
tyrosylprotein sulfotransferase 1, like |
| chr6_-_28117995 | 0.61 |
ENSDART00000147253
|
si:ch73-194h10.3
|
si:ch73-194h10.3 |
| chr10_+_1849874 | 0.61 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr20_-_154989 | 0.60 |
ENSDART00000064542
|
rpf2
|
ribosome production factor 2 homolog |
| chr21_-_1640547 | 0.60 |
ENSDART00000151041
|
zgc:152948
|
zgc:152948 |
| chr9_-_50000144 | 0.60 |
ENSDART00000123416
|
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr3_-_55531450 | 0.60 |
ENSDART00000155376
|
tex2
|
testis expressed 2 |
| chr22_+_10163901 | 0.60 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr24_+_31459715 | 0.59 |
ENSDART00000181102
ENSDART00000189950 ENSDART00000192321 ENSDART00000126380 |
cnbd1
|
cyclic nucleotide binding domain containing 1 |
| chr17_+_6538733 | 0.58 |
ENSDART00000193712
|
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr2_-_37744951 | 0.57 |
ENSDART00000144807
|
myo9b
|
myosin IXb |
| chr19_-_79202 | 0.57 |
ENSDART00000166009
|
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr3_+_40289418 | 0.57 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr2_-_56649883 | 0.56 |
ENSDART00000191786
|
gpx4b
|
glutathione peroxidase 4b |
| chr11_+_25403561 | 0.56 |
ENSDART00000089120
|
adnpa
|
activity-dependent neuroprotector homeobox a |
| chr17_-_43594864 | 0.56 |
ENSDART00000139980
|
zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr18_-_35736591 | 0.56 |
ENSDART00000036015
|
ryr1b
|
ryanodine receptor 1b (skeletal) |
| chr1_-_51720633 | 0.55 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr6_-_41138854 | 0.54 |
ENSDART00000128723
ENSDART00000151055 ENSDART00000132484 |
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr8_-_43716897 | 0.54 |
ENSDART00000163237
|
ep400
|
E1A binding protein p400 |
| chr21_-_1972236 | 0.54 |
ENSDART00000192858
ENSDART00000189962 ENSDART00000182461 ENSDART00000165547 |
WDR7
|
WD repeat domain 7 |
| chr10_+_45128375 | 0.54 |
ENSDART00000164805
|
camk2b2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II beta 2 |
| chr20_+_13969414 | 0.53 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr15_+_40074923 | 0.53 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr10_-_35542071 | 0.53 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr4_-_14531687 | 0.52 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr6_-_11759860 | 0.51 |
ENSDART00000151296
|
si:ch211-162i14.1
|
si:ch211-162i14.1 |
| chr19_+_32158010 | 0.51 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr2_-_32688905 | 0.51 |
ENSDART00000041146
|
nrbp2a
|
nuclear receptor binding protein 2a |
| chr20_-_7128612 | 0.50 |
ENSDART00000146755
ENSDART00000036871 |
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr18_+_38807239 | 0.49 |
ENSDART00000184332
|
fam214a
|
family with sequence similarity 214, member A |
| chr2_-_10386738 | 0.49 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr20_-_42241456 | 0.48 |
ENSDART00000034054
|
nus1
|
NUS1 dehydrodolichyl diphosphate synthase subunit |
| chr15_+_47618221 | 0.48 |
ENSDART00000168722
|
paf1
|
PAF1 homolog, Paf1/RNA polymerase II complex component |
| chr22_-_11833317 | 0.48 |
ENSDART00000125423
ENSDART00000000192 |
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr21_+_15704556 | 0.47 |
ENSDART00000024858
ENSDART00000146909 |
chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10 |
| chr2_-_42558549 | 0.47 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr7_+_38588866 | 0.47 |
ENSDART00000015682
|
ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3, (NADH-coenzyme Q reductase) |
| chr2_+_26240631 | 0.47 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr25_+_22017182 | 0.46 |
ENSDART00000156517
|
si:dkey-217l24.1
|
si:dkey-217l24.1 |
| chr7_+_69019851 | 0.45 |
ENSDART00000162891
|
CABZ01057488.1
|
|
| chr16_+_7991274 | 0.44 |
ENSDART00000179704
|
ano10a
|
anoctamin 10a |
| chr25_-_25575241 | 0.44 |
ENSDART00000150636
|
hic1l
|
hypermethylated in cancer 1 like |
| chr20_+_47953047 | 0.43 |
ENSDART00000079734
|
hadhaa
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha a |
| chr18_-_12957451 | 0.43 |
ENSDART00000140403
|
srgap1a
|
SLIT-ROBO Rho GTPase activating protein 1a |
| chr9_+_54686686 | 0.43 |
ENSDART00000066198
|
rab9a
|
RAB9A, member RAS oncogene family |
| chr23_-_10696626 | 0.43 |
ENSDART00000177571
|
foxp1a
|
forkhead box P1a |
| chr5_+_6670945 | 0.42 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr23_-_24234371 | 0.42 |
ENSDART00000124539
|
ddost
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr2_-_15362923 | 0.42 |
ENSDART00000166926
|
olfm3b
|
olfactomedin 3b |
| chr2_+_3823813 | 0.42 |
ENSDART00000103596
ENSDART00000161880 ENSDART00000185408 |
npc1
|
Niemann-Pick disease, type C1 |
| chr13_-_8692432 | 0.41 |
ENSDART00000058106
|
mcfd2
|
multiple coagulation factor deficiency 2 |
| chr20_-_48604199 | 0.41 |
ENSDART00000161762
ENSDART00000170894 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr2_-_39017838 | 0.41 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr4_-_858434 | 0.41 |
ENSDART00000006961
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr8_+_32742650 | 0.41 |
ENSDART00000138117
|
hmcn2
|
hemicentin 2 |
| chr3_+_16922226 | 0.41 |
ENSDART00000017646
|
atp6v0a1a
|
ATPase H+ transporting V0 subunit a1a |
| chr13_-_35808904 | 0.40 |
ENSDART00000171667
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr8_+_19624589 | 0.39 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr22_+_4447387 | 0.39 |
ENSDART00000189742
ENSDART00000166768 |
timm44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr11_-_44999858 | 0.39 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr1_-_44581937 | 0.39 |
ENSDART00000009858
|
tmx2b
|
thioredoxin-related transmembrane protein 2b |
| chr10_+_20180163 | 0.38 |
ENSDART00000080016
|
ppp3ccb
|
protein phosphatase 3, catalytic subunit, gamma isozyme, b |
| chr10_+_44692272 | 0.38 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr17_+_33418475 | 0.37 |
ENSDART00000169145
|
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr1_-_46859398 | 0.37 |
ENSDART00000135795
|
agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr16_-_53800047 | 0.37 |
ENSDART00000158047
|
znrf2b
|
zinc and ring finger 2b |
| chr13_-_42724645 | 0.36 |
ENSDART00000046066
|
capn1a
|
calpain 1, (mu/I) large subunit a |
| chr6_-_14004772 | 0.36 |
ENSDART00000185629
|
zgc:92027
|
zgc:92027 |
| chr11_-_27778831 | 0.36 |
ENSDART00000023823
|
cass4
|
Cas scaffolding protein family member 4 |
| chr19_+_41701660 | 0.36 |
ENSDART00000033362
|
gatad2b
|
GATA zinc finger domain containing 2B |
| chr2_-_37277626 | 0.35 |
ENSDART00000135340
|
nadkb
|
NAD kinase b |
| chr9_+_17429170 | 0.35 |
ENSDART00000006256
|
zgc:101559
|
zgc:101559 |
| chr7_-_51461649 | 0.34 |
ENSDART00000193947
ENSDART00000174328 |
arhgap36
|
Rho GTPase activating protein 36 |
| chr17_-_25303486 | 0.34 |
ENSDART00000162235
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr24_+_19522094 | 0.34 |
ENSDART00000191042
ENSDART00000184714 ENSDART00000180782 |
sulf1
|
sulfatase 1 |
| chr22_-_15720347 | 0.33 |
ENSDART00000105692
|
mrpl54
|
mitochondrial ribosomal protein L54 |
| chr19_-_24136233 | 0.33 |
ENSDART00000143365
|
thap7
|
THAP domain containing 7 |
| chr16_+_33163858 | 0.33 |
ENSDART00000101943
|
rragca
|
Ras-related GTP binding Ca |
| chr6_-_21830405 | 0.33 |
ENSDART00000151803
ENSDART00000113497 |
setd5
|
SET domain containing 5 |
| chr17_+_450956 | 0.33 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr10_+_31244619 | 0.33 |
ENSDART00000145562
ENSDART00000184412 |
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr18_+_25546227 | 0.33 |
ENSDART00000085824
|
pex11a
|
peroxisomal biogenesis factor 11 alpha |
| chr6_-_35046735 | 0.33 |
ENSDART00000143649
|
uap1
|
UDP-N-acetylglucosamine pyrophosphorylase 1 |
| chr7_+_65398161 | 0.32 |
ENSDART00000166109
ENSDART00000157399 |
usp47
|
ubiquitin specific peptidase 47 |
| chr21_+_1119046 | 0.32 |
ENSDART00000184678
|
CABZ01088049.1
|
|
| chr21_-_13055195 | 0.32 |
ENSDART00000133517
|
myorg
|
myogenesis regulating glycosidase (putative) |
| chr5_+_63329608 | 0.32 |
ENSDART00000139180
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr15_-_16946124 | 0.32 |
ENSDART00000154923
|
hip1
|
huntingtin interacting protein 1 |
| chr21_-_30166097 | 0.31 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr20_-_29505863 | 0.31 |
ENSDART00000148278
|
klf11a
|
Kruppel-like factor 11a |
| chr12_+_1286642 | 0.31 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr15_+_24756860 | 0.31 |
ENSDART00000156424
ENSDART00000078035 |
cpda
|
carboxypeptidase D, a |
| chr20_+_6535438 | 0.30 |
ENSDART00000145763
|
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr21_+_15883546 | 0.30 |
ENSDART00000186325
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr20_-_48604621 | 0.29 |
ENSDART00000161769
ENSDART00000157871 |
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr5_-_37881345 | 0.29 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr23_-_1587955 | 0.29 |
ENSDART00000136037
|
fndc7b
|
fibronectin type III domain containing 7b |
| chr17_+_15213496 | 0.29 |
ENSDART00000058351
ENSDART00000131663 |
gnpnat1
|
glucosamine-phosphate N-acetyltransferase 1 |
| chr22_-_14262115 | 0.29 |
ENSDART00000168264
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr23_-_23179417 | 0.28 |
ENSDART00000122945
ENSDART00000091662 |
noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr2_+_16460823 | 0.28 |
ENSDART00000146855
|
agfg1b
|
ArfGAP with FG repeats 1b |
| chr21_-_4250682 | 0.28 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr12_-_47774807 | 0.28 |
ENSDART00000193831
|
LO017725.1
|
|
| chr11_+_22134540 | 0.28 |
ENSDART00000190502
|
MDFI
|
si:dkey-91m3.1 |
| chr13_-_30142087 | 0.27 |
ENSDART00000110157
|
tysnd1
|
trypsin domain containing 1 |
| chr7_+_34296789 | 0.27 |
ENSDART00000052471
ENSDART00000173798 ENSDART00000173778 |
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr6_-_7082108 | 0.27 |
ENSDART00000081760
|
ihhb
|
Indian hedgehog homolog b |
| chr12_+_26670778 | 0.27 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr8_-_65189 | 0.27 |
ENSDART00000168412
|
hsd17b4
|
hydroxysteroid (17-beta) dehydrogenase 4 |
| chr23_-_46126444 | 0.26 |
ENSDART00000030004
|
CABZ01071903.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of rxraa+rxrab+rxrba+rxrgb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0050787 | response to mercury ion(GO:0046689) detoxification of mercury ion(GO:0050787) |
| 0.5 | 2.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.2 | GO:0019408 | dolichol biosynthetic process(GO:0019408) |
| 0.4 | 1.2 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.4 | 1.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.3 | 1.0 | GO:0042706 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.2 | 2.0 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 | 0.7 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 | 0.9 | GO:0006549 | isoleucine metabolic process(GO:0006549) isoleucine biosynthetic process(GO:0009097) |
| 0.2 | 1.2 | GO:0051791 | medium-chain fatty acid metabolic process(GO:0051791) medium-chain fatty acid catabolic process(GO:0051793) |
| 0.2 | 0.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.2 | 0.7 | GO:2000623 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.2 | 1.6 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.2 | 0.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.6 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.6 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.1 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 0.7 | GO:0033539 | carnitine metabolic process, CoA-linked(GO:0019254) fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.7 | GO:1901842 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 1.0 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.1 | 6.7 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.4 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 | 0.7 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.6 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.1 | 5.6 | GO:0048599 | oocyte development(GO:0048599) |
| 0.1 | 0.3 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.1 | 0.9 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 2.3 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 0.6 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 1.5 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 9.3 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.1 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.1 | 0.6 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.2 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.7 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 0.6 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.4 | GO:0043589 | skin morphogenesis(GO:0043589) mesenchymal cell migration(GO:0090497) |
| 0.1 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.1 | 0.9 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.8 | GO:0061055 | myotome development(GO:0061055) |
| 0.1 | 0.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.6 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.3 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.3 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 1.4 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.2 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.7 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.3 | GO:0007176 | regulation of epidermal growth factor-activated receptor activity(GO:0007176) |
| 0.0 | 0.5 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:1905048 | regulation of metallopeptidase activity(GO:1905048) |
| 0.0 | 1.1 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 0.4 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.8 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0035065 | regulation of histone acetylation(GO:0035065) |
| 0.0 | 1.2 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.0 | 0.4 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.8 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.2 | GO:0051896 | regulation of protein kinase B signaling(GO:0051896) |
| 0.0 | 0.3 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.1 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.4 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.6 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 1.0 | GO:0000723 | telomere maintenance(GO:0000723) telomere organization(GO:0032200) |
| 0.0 | 0.0 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.2 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.5 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.6 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.1 | GO:0048660 | regulation of smooth muscle cell proliferation(GO:0048660) negative regulation of smooth muscle cell proliferation(GO:0048662) |
| 0.0 | 0.6 | GO:0007006 | mitochondrial membrane organization(GO:0007006) |
| 0.0 | 0.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.3 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.3 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.5 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.0 | GO:0072020 | proximal straight tubule development(GO:0072020) |
| 0.0 | 0.2 | GO:0007525 | somatic muscle development(GO:0007525) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 0.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.4 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.0 | GO:0015884 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.1 | GO:0033700 | phospholipid efflux(GO:0033700) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.4 | 1.1 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.3 | 2.3 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.3 | 1.2 | GO:1904423 | dehydrodolichyl diphosphate synthase complex(GO:1904423) |
| 0.3 | 0.9 | GO:0005948 | acetolactate synthase complex(GO:0005948) |
| 0.3 | 0.8 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.2 | 0.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 0.6 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 0.6 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.5 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 1.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 0.6 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 2.0 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 3.2 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.6 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.3 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.6 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.0 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 0.1 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.1 | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.1 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.5 | 2.3 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.4 | 6.7 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.3 | 1.2 | GO:0045547 | dehydrodolichyl diphosphate synthase activity(GO:0045547) |
| 0.3 | 0.9 | GO:0003984 | acetolactate synthase activity(GO:0003984) |
| 0.3 | 2.3 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.2 | 0.7 | GO:1990825 | mRNA CDS binding(GO:1990715) sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 0.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.2 | 0.6 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.2 | 1.2 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.8 | GO:0008469 | histone-arginine N-methyltransferase activity(GO:0008469) |
| 0.1 | 0.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 0.7 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.6 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 0.6 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.1 | 1.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.6 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 0.2 | GO:0043878 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) |
| 0.1 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.3 | GO:0003977 | UDP-N-acetylglucosamine diphosphorylase activity(GO:0003977) |
| 0.1 | 0.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.2 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.1 | 0.2 | GO:1990931 | oxidative DNA demethylase activity(GO:0035516) DNA-N1-methyladenine dioxygenase activity(GO:0043734) RNA N6-methyladenosine dioxygenase activity(GO:1990931) |
| 0.0 | 0.6 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.7 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.4 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.5 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 8.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.8 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.1 | GO:0004557 | alpha-galactosidase activity(GO:0004557) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.0 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 2.4 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.2 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.7 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.0 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.5 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.5 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0099589 | serotonin receptor activity(GO:0099589) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.4 | GO:0043492 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 0.8 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.0 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.4 | GO:0015036 | disulfide oxidoreductase activity(GO:0015036) |
| 0.0 | 0.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 2.0 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 1.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 1.1 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |