Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
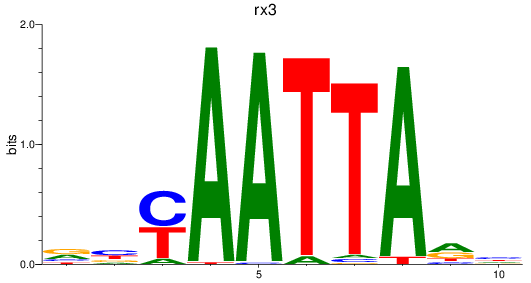
Results for rx3
Z-value: 1.06
Transcription factors associated with rx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rx3
|
ENSDARG00000052893 | retinal homeobox gene 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx3 | dr11_v1_chr21_+_10756154_10756154 | -0.89 | 7.4e-07 | Click! |
Activity profile of rx3 motif
Sorted Z-values of rx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_21362071 | 4.64 |
ENSDART00000125167
|
avd
|
avidin |
| chr10_-_21362320 | 4.35 |
ENSDART00000189789
|
avd
|
avidin |
| chr9_-_35633827 | 4.26 |
ENSDART00000077745
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr20_-_23426339 | 4.13 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr18_-_40708537 | 4.00 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr21_+_25777425 | 3.60 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr8_+_45334255 | 3.42 |
ENSDART00000126848
ENSDART00000134161 ENSDART00000142322 ENSDART00000145011 ENSDART00000183560 |
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr11_-_44801968 | 3.22 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr1_-_18811517 | 3.05 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr10_-_34002185 | 3.02 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr2_+_6253246 | 2.95 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr17_+_16046314 | 2.12 |
ENSDART00000154554
ENSDART00000154338 ENSDART00000155336 |
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr12_-_33357655 | 2.11 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr2_-_15324837 | 1.93 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr6_+_21001264 | 1.91 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr17_+_16046132 | 1.90 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr24_+_12835935 | 1.73 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr17_-_40956035 | 1.66 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr21_-_32060993 | 1.36 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr1_-_513762 | 1.35 |
ENSDART00000148162
ENSDART00000144606 |
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr14_+_34490445 | 1.29 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr20_-_40758410 | 1.20 |
ENSDART00000183031
|
cx34.5
|
connexin 34.5 |
| chr19_-_25119443 | 1.16 |
ENSDART00000148953
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_-_19467011 | 1.09 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr12_+_22580579 | 1.06 |
ENSDART00000171725
ENSDART00000192290 |
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr13_+_38814521 | 1.03 |
ENSDART00000110976
|
col19a1
|
collagen, type XIX, alpha 1 |
| chr19_-_8768564 | 1.02 |
ENSDART00000170416
|
si:ch73-350k19.1
|
si:ch73-350k19.1 |
| chr10_+_43039947 | 1.00 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr14_+_8940326 | 0.98 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr17_+_37227936 | 0.95 |
ENSDART00000076009
|
hadhab
|
hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b |
| chr8_-_25034411 | 0.94 |
ENSDART00000135973
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr6_+_40922572 | 0.93 |
ENSDART00000133599
ENSDART00000002728 ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr5_+_60590796 | 0.88 |
ENSDART00000159859
|
tmem132e
|
transmembrane protein 132E |
| chr17_+_19630272 | 0.87 |
ENSDART00000104895
|
rgs7a
|
regulator of G protein signaling 7a |
| chr24_-_25144441 | 0.85 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr1_+_35985813 | 0.84 |
ENSDART00000179634
ENSDART00000139636 ENSDART00000175902 |
zgc:152968
|
zgc:152968 |
| chr20_-_37813863 | 0.79 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr25_-_27621268 | 0.78 |
ENSDART00000146205
ENSDART00000073511 |
hyal6
|
hyaluronoglucosaminidase 6 |
| chr6_-_12172424 | 0.77 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr10_-_32494499 | 0.70 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr14_+_30795559 | 0.70 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr9_+_43799829 | 0.69 |
ENSDART00000186240
|
ube2e3
|
ubiquitin-conjugating enzyme E2E 3 (UBC4/5 homolog, yeast) |
| chr1_+_513986 | 0.69 |
ENSDART00000109083
ENSDART00000081945 |
txnl4b
|
thioredoxin-like 4B |
| chr6_-_40922971 | 0.67 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr22_-_21897203 | 0.66 |
ENSDART00000158501
ENSDART00000105566 ENSDART00000136795 |
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr17_+_24821627 | 0.66 |
ENSDART00000112389
|
wdr43
|
WD repeat domain 43 |
| chr16_-_42056137 | 0.66 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr24_+_19415124 | 0.65 |
ENSDART00000186931
|
sulf1
|
sulfatase 1 |
| chr2_-_21820697 | 0.59 |
ENSDART00000135230
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr10_-_32494304 | 0.59 |
ENSDART00000028161
|
uvrag
|
UV radiation resistance associated gene |
| chr7_-_30174882 | 0.57 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr13_+_22480496 | 0.55 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr4_-_4834617 | 0.53 |
ENSDART00000141539
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr13_+_22479988 | 0.53 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr2_+_40294313 | 0.52 |
ENSDART00000037292
|
epha4b
|
eph receptor A4b |
| chr9_-_3934963 | 0.52 |
ENSDART00000062336
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr23_-_17003533 | 0.50 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr9_+_50001746 | 0.50 |
ENSDART00000058892
|
slc38a11
|
solute carrier family 38, member 11 |
| chr4_-_4834347 | 0.48 |
ENSDART00000141803
|
coa6
|
cytochrome c oxidase assembly factor 6 |
| chr24_-_39518599 | 0.47 |
ENSDART00000145606
ENSDART00000031486 |
lyrm1
|
LYR motif containing 1 |
| chr20_+_28803977 | 0.46 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr17_+_8799661 | 0.45 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr8_+_25034544 | 0.45 |
ENSDART00000123300
|
ngrn
|
neugrin, neurite outgrowth associated |
| chr9_-_50001606 | 0.44 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr10_-_13343831 | 0.42 |
ENSDART00000135941
|
il11ra
|
interleukin 11 receptor, alpha |
| chr15_-_9272328 | 0.42 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr2_+_50608099 | 0.40 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr13_-_31017960 | 0.38 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr20_-_9436521 | 0.38 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr16_+_27383717 | 0.37 |
ENSDART00000132329
ENSDART00000136256 |
stx17
|
syntaxin 17 |
| chr22_-_20166660 | 0.37 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr17_+_8799451 | 0.36 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr8_-_50888806 | 0.34 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr20_-_45060241 | 0.33 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr24_-_25004553 | 0.33 |
ENSDART00000080997
ENSDART00000136860 |
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr3_-_61162750 | 0.33 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr10_-_33297864 | 0.26 |
ENSDART00000163360
|
PRDM15
|
PR/SET domain 15 |
| chr1_-_26444075 | 0.26 |
ENSDART00000125690
|
ints12
|
integrator complex subunit 12 |
| chr4_+_3980247 | 0.26 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr13_+_38430466 | 0.24 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr3_+_32365811 | 0.23 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr22_-_7129631 | 0.23 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr11_+_31864921 | 0.21 |
ENSDART00000180252
|
diaph3
|
diaphanous-related formin 3 |
| chr15_+_31344472 | 0.21 |
ENSDART00000146695
ENSDART00000159182 ENSDART00000060125 |
or107-1
|
odorant receptor, family D, subfamily 107, member 1 |
| chr15_-_2184638 | 0.21 |
ENSDART00000135460
|
shox2
|
short stature homeobox 2 |
| chr9_+_22485343 | 0.20 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr14_+_36521005 | 0.17 |
ENSDART00000192286
|
TENM3
|
si:dkey-237h12.3 |
| chr24_+_39518774 | 0.15 |
ENSDART00000132939
|
dcun1d3
|
defective in cullin neddylation 1 domain containing 3 |
| chr16_+_42471455 | 0.15 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr3_+_23692462 | 0.14 |
ENSDART00000145934
|
hoxb7a
|
homeobox B7a |
| chr11_+_33312601 | 0.14 |
ENSDART00000188024
|
cntnap5l
|
contactin associated protein-like 5 like |
| chr22_+_28337204 | 0.14 |
ENSDART00000163352
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr12_+_48803098 | 0.14 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr13_-_25819825 | 0.13 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr15_-_5178899 | 0.12 |
ENSDART00000132148
|
or126-4
|
odorant receptor, family E, subfamily 126, member 4 |
| chr8_+_7801060 | 0.11 |
ENSDART00000161618
|
tfe3a
|
transcription factor binding to IGHM enhancer 3a |
| chr14_-_7207961 | 0.11 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr23_-_1348933 | 0.11 |
ENSDART00000168981
|
CABZ01078120.1
|
|
| chr8_+_6410933 | 0.11 |
ENSDART00000168789
|
lrrtm4l2
|
leucine rich repeat transmembrane neuronal 4 like 2 |
| chr21_-_15200556 | 0.10 |
ENSDART00000141809
|
sfswap
|
splicing factor SWAP |
| chr22_+_28337429 | 0.10 |
ENSDART00000166177
|
impg2b
|
interphotoreceptor matrix proteoglycan 2b |
| chr3_-_50443607 | 0.10 |
ENSDART00000074036
|
rcvrna
|
recoverin a |
| chr7_-_23768234 | 0.09 |
ENSDART00000173981
|
si:ch211-200p22.4
|
si:ch211-200p22.4 |
| chr19_+_2631565 | 0.08 |
ENSDART00000171487
|
fam126a
|
family with sequence similarity 126, member A |
| chr17_-_200316 | 0.08 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr20_-_29864390 | 0.08 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr7_+_34592526 | 0.08 |
ENSDART00000173959
|
fhod1
|
formin homology 2 domain containing 1 |
| chr8_-_39822917 | 0.06 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr18_-_5598958 | 0.06 |
ENSDART00000161538
|
cyp1a
|
cytochrome P450, family 1, subfamily A |
| chr19_-_30524952 | 0.06 |
ENSDART00000103506
|
hpcal4
|
hippocalcin like 4 |
| chr11_-_29563437 | 0.05 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr21_+_28478663 | 0.04 |
ENSDART00000077887
ENSDART00000134150 |
slc22a6l
|
solute carrier family 22 (organic anion transporter), member 6, like |
| chr15_-_16177603 | 0.02 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr17_-_20717845 | 0.01 |
ENSDART00000150037
|
ank3b
|
ankyrin 3b |
| chr14_-_32016615 | 0.00 |
ENSDART00000105761
|
zic3
|
zic family member 3 heterotaxy 1 (odd-paired homolog, Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of rx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.4 | 1.3 | GO:0072111 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.3 | 1.3 | GO:0090646 | mitochondrial tRNA processing(GO:0090646) |
| 0.2 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 3.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.0 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.2 | 3.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.5 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.5 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 1.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.7 | GO:2000290 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) regulation of myotome development(GO:2000290) |
| 0.1 | 0.9 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.1 | 0.7 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.8 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 1.9 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.1 | 0.4 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 1.0 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.6 | GO:0009303 | rRNA transcription(GO:0009303) swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 5.1 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 0.8 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.3 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 1.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.4 | GO:0060968 | regulation of gene silencing(GO:0060968) |
| 0.0 | 0.2 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 1.0 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 1.6 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 1.3 | GO:0006497 | protein lipidation(GO:0006497) |
| 0.0 | 0.7 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0051443 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.2 | 1.3 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 0.5 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 3.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 2.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.7 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.9 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 3.2 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.1 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 4.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 9.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 1.3 | GO:0052905 | tRNA (guanine(9)-N(1))-methyltransferase activity(GO:0052905) |
| 0.3 | 1.0 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.3 | 1.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.3 | 2.9 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.2 | 3.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 2.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.5 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.1 | 0.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 0.8 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.1 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 3.2 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.7 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.9 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 7.4 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 5.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.7 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.2 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |