Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
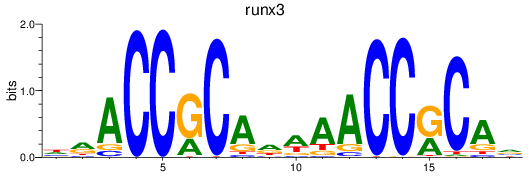
Results for runx3
Z-value: 0.41
Transcription factors associated with runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx3
|
ENSDARG00000052826 | RUNX family transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx3 | dr11_v1_chr13_-_45123051_45123051 | 0.92 | 4.1e-08 | Click! |
Activity profile of runx3 motif
Sorted Z-values of runx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_2642013 | 0.59 |
ENSDART00000111324
|
zgc:193807
|
zgc:193807 |
| chr7_+_24888910 | 0.52 |
ENSDART00000173994
|
mark2b
|
MAP/microtubule affinity-regulating kinase 2b |
| chr22_-_25469751 | 0.52 |
ENSDART00000171670
|
CR769772.4
|
|
| chr18_+_35173859 | 0.52 |
ENSDART00000127379
ENSDART00000098292 |
cfap45
|
cilia and flagella associated protein 45 |
| chr18_+_35173683 | 0.50 |
ENSDART00000192545
|
cfap45
|
cilia and flagella associated protein 45 |
| chr20_-_43346049 | 0.50 |
ENSDART00000152888
|
afdna
|
afadin, adherens junction formation factor a |
| chr22_-_25502977 | 0.48 |
ENSDART00000181749
|
CR769772.4
|
|
| chr19_-_5380770 | 0.46 |
ENSDART00000000221
|
krt91
|
keratin 91 |
| chr16_+_23431189 | 0.43 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr16_+_41060161 | 0.38 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr5_+_24059598 | 0.36 |
ENSDART00000051547
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr11_-_30636163 | 0.35 |
ENSDART00000140516
|
zgc:153665
|
zgc:153665 |
| chr11_-_8208464 | 0.35 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr6_+_44163727 | 0.35 |
ENSDART00000064878
|
gxylt2
|
glucoside xylosyltransferase 2 |
| chr3_-_3372259 | 0.34 |
ENSDART00000140482
|
si:dkey-46g23.1
|
si:dkey-46g23.1 |
| chr21_+_11923701 | 0.33 |
ENSDART00000109292
|
ubap2a
|
ubiquitin associated protein 2a |
| chr17_+_7522777 | 0.32 |
ENSDART00000184723
|
klhl10b.2
|
kelch-like family member 10b, tandem duplicate 2 |
| chr15_-_2657508 | 0.30 |
ENSDART00000102086
|
cldna
|
claudin a |
| chr23_+_20795781 | 0.29 |
ENSDART00000128577
|
ttc34
|
tetratricopeptide repeat domain 34 |
| chr24_+_24064562 | 0.28 |
ENSDART00000144394
|
zgc:112408
|
zgc:112408 |
| chr16_+_28754403 | 0.25 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr25_-_10503043 | 0.25 |
ENSDART00000155404
|
cox8b
|
cytochrome c oxidase subunit 8b |
| chr20_-_34670236 | 0.24 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr7_+_41160225 | 0.23 |
ENSDART00000074212
ENSDART00000164200 ENSDART00000174204 ENSDART00000174212 |
scrib
|
scribbled planar cell polarity protein |
| chr2_+_49864219 | 0.22 |
ENSDART00000187744
|
rpl37
|
ribosomal protein L37 |
| chr1_+_54911458 | 0.22 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chr20_-_14012859 | 0.21 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr3_-_49382896 | 0.21 |
ENSDART00000169115
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr1_+_46598764 | 0.21 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr1_+_53321878 | 0.21 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr9_-_20372977 | 0.20 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr7_+_25858380 | 0.20 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr14_-_42231293 | 0.19 |
ENSDART00000185486
|
BX890543.1
|
|
| chr5_+_7989210 | 0.19 |
ENSDART00000168071
|
gdnfb
|
glial cell derived neurotrophic factor b |
| chr19_-_72398 | 0.19 |
ENSDART00000165587
|
dnajc8
|
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chr15_-_31067589 | 0.18 |
ENSDART00000060157
|
lgals9l3
|
lectin, galactoside-binding, soluble, 9 (galectin 9)-like 3 |
| chr2_-_183992 | 0.18 |
ENSDART00000126704
ENSDART00000191283 ENSDART00000034783 |
zgc:113518
|
zgc:113518 |
| chr19_+_2546775 | 0.17 |
ENSDART00000148527
ENSDART00000097528 |
sp4
|
sp4 transcription factor |
| chr16_+_50089417 | 0.17 |
ENSDART00000153675
|
nr1d2a
|
nuclear receptor subfamily 1, group D, member 2a |
| chr20_-_2725930 | 0.17 |
ENSDART00000081643
|
akirin2
|
akirin 2 |
| chr22_-_14161309 | 0.16 |
ENSDART00000133365
|
si:ch211-246m6.5
|
si:ch211-246m6.5 |
| chr18_-_50524017 | 0.16 |
ENSDART00000150013
ENSDART00000149912 |
cd276
|
CD276 molecule |
| chr18_-_50523399 | 0.15 |
ENSDART00000033591
|
cd276
|
CD276 molecule |
| chr2_+_4146606 | 0.15 |
ENSDART00000171170
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr18_-_7810214 | 0.15 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr25_-_225964 | 0.15 |
ENSDART00000193424
|
CABZ01113818.1
|
|
| chr25_-_21894317 | 0.14 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| chr15_-_20468302 | 0.13 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr7_+_17937922 | 0.13 |
ENSDART00000147189
|
mta2
|
metastasis associated 1 family, member 2 |
| chr2_+_16173999 | 0.13 |
ENSDART00000177639
ENSDART00000157601 ENSDART00000190186 ENSDART00000045933 |
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr5_-_10239079 | 0.13 |
ENSDART00000132739
|
si:ch73-42k18.1
|
si:ch73-42k18.1 |
| chr22_+_9922301 | 0.12 |
ENSDART00000105924
|
blf
|
bloody fingers |
| chr7_+_17938128 | 0.12 |
ENSDART00000141044
|
mta2
|
metastasis associated 1 family, member 2 |
| chr16_-_54907588 | 0.12 |
ENSDART00000185709
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr22_+_11520249 | 0.11 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr22_-_30973791 | 0.11 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr8_-_18899427 | 0.10 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr13_+_45524475 | 0.09 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr11_+_30636351 | 0.09 |
ENSDART00000087909
|
tmem246
|
transmembrane protein 246 |
| chr24_-_21507627 | 0.09 |
ENSDART00000170984
|
rnf6
|
ring finger protein (C3H2C3 type) 6 |
| chr15_-_37589600 | 0.07 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr16_+_4133519 | 0.07 |
ENSDART00000174521
ENSDART00000175718 |
mtf1
|
metal-regulatory transcription factor 1 |
| chr2_-_52841762 | 0.07 |
ENSDART00000114682
|
ralbp1
|
ralA binding protein 1 |
| chr2_+_53114476 | 0.07 |
ENSDART00000080850
ENSDART00000104187 |
CR384061.1
|
|
| chr6_-_2154137 | 0.07 |
ENSDART00000162656
|
tgm5l
|
transglutaminase 5, like |
| chr15_-_22139566 | 0.07 |
ENSDART00000149017
|
scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr20_-_48485354 | 0.06 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr20_+_10723292 | 0.06 |
ENSDART00000152805
|
si:ch211-182e10.4
|
si:ch211-182e10.4 |
| chr6_-_141564 | 0.06 |
ENSDART00000151245
ENSDART00000063876 |
s1pr5b
|
sphingosine-1-phosphate receptor 5b |
| chr1_-_18446161 | 0.06 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr6_+_39930825 | 0.05 |
ENSDART00000182352
ENSDART00000186643 ENSDART00000190513 ENSDART00000065092 ENSDART00000188645 |
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr20_-_53321499 | 0.05 |
ENSDART00000179894
ENSDART00000127427 ENSDART00000084952 |
wasf1
|
WAS protein family, member 1 |
| chr25_-_210730 | 0.05 |
ENSDART00000187580
|
FP236318.1
|
|
| chr2_-_7246848 | 0.05 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr24_-_18659147 | 0.05 |
ENSDART00000166039
|
zgc:66014
|
zgc:66014 |
| chr11_-_40519886 | 0.05 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr19_-_24443867 | 0.04 |
ENSDART00000163763
ENSDART00000043133 |
thbs3b
|
thrombospondin 3b |
| chr22_-_392224 | 0.04 |
ENSDART00000124720
|
CABZ01072989.1
|
|
| chr6_-_9581949 | 0.04 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr7_-_65492048 | 0.03 |
ENSDART00000162381
|
dkk3a
|
dickkopf WNT signaling pathway inhibitor 3a |
| chr10_+_45148167 | 0.03 |
ENSDART00000172621
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr8_+_24861264 | 0.03 |
ENSDART00000099607
|
slc6a17
|
solute carrier family 6 (neutral amino acid transporter), member 17 |
| chr20_+_44498056 | 0.02 |
ENSDART00000023763
|
wdcp
|
WD repeat and coiled coil containing |
| chr21_+_15824182 | 0.02 |
ENSDART00000065779
|
gnrh2
|
gonadotropin-releasing hormone 2 |
| chr7_+_73308566 | 0.02 |
ENSDART00000187039
ENSDART00000174244 |
CABZ01081777.1
|
|
| chr22_+_10757848 | 0.01 |
ENSDART00000145551
|
tmprss9
|
transmembrane protease, serine 9 |
| chr1_-_8012476 | 0.01 |
ENSDART00000177051
|
CR855320.3
|
|
| chr22_+_18497759 | 0.01 |
ENSDART00000188973
|
BX649279.1
|
|
| chr19_+_42219165 | 0.01 |
ENSDART00000163192
|
CU896644.1
|
|
| chr10_+_45148005 | 0.01 |
ENSDART00000182501
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr21_-_43992027 | 0.01 |
ENSDART00000188612
|
cdx1b
|
caudal type homeobox 1 b |
| chr17_+_37310663 | 0.00 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr22_+_11144153 | 0.00 |
ENSDART00000047442
|
bcor
|
BCL6 corepressor |
| chr24_-_27423697 | 0.00 |
ENSDART00000132590
|
si:dkey-25o1.7
|
si:dkey-25o1.7 |
| chr6_+_38896158 | 0.00 |
ENSDART00000029930
ENSDART00000131347 |
slc48a1b
|
solute carrier family 48 (heme transporter), member 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061213 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.1 | 0.2 | GO:0021527 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.3 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.2 | GO:2000561 | negative regulation of interferon-gamma production(GO:0032689) CD4-positive, alpha-beta T cell proliferation(GO:0035739) regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000561) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.4 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.3 | GO:0040033 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0071276 | cellular response to cadmium ion(GO:0071276) |
| 0.0 | 0.1 | GO:0086005 | ventricular cardiac muscle cell action potential(GO:0086005) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) |
| 0.0 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |