Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
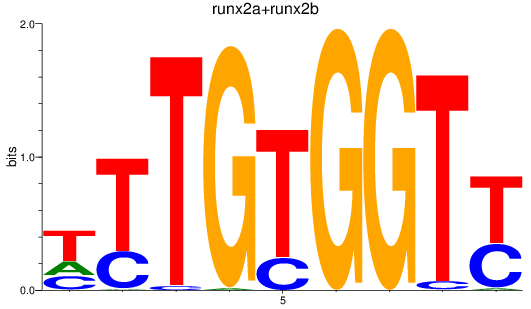
Results for runx2a+runx2b
Z-value: 0.83
Transcription factors associated with runx2a+runx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx2a
|
ENSDARG00000040261 | RUNX family transcription factor 2a |
|
runx2b
|
ENSDARG00000059233 | RUNX family transcription factor 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx2a | dr11_v1_chr17_+_5351922_5351922 | -0.32 | 2.0e-01 | Click! |
| runx2b | dr11_v1_chr20_-_44055095_44055123 | 0.25 | 3.3e-01 | Click! |
Activity profile of runx2a+runx2b motif
Sorted Z-values of runx2a+runx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_24488652 | 2.52 |
ENSDART00000052067
|
insl3
|
insulin-like 3 (Leydig cell) |
| chr24_+_31361407 | 1.79 |
ENSDART00000162668
|
cremb
|
cAMP responsive element modulator b |
| chr24_+_17269849 | 1.75 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr17_+_21295132 | 1.74 |
ENSDART00000103845
|
eno4
|
enolase family member 4 |
| chr16_+_28727383 | 1.56 |
ENSDART00000149753
|
dcst2
|
DC-STAMP domain containing 2 |
| chr17_+_26815021 | 1.48 |
ENSDART00000086885
|
asmt2
|
acetylserotonin O-methyltransferase 2 |
| chr5_+_13359146 | 1.38 |
ENSDART00000179839
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr22_-_36934040 | 1.36 |
ENSDART00000151666
|
pimr206
|
Pim proto-oncogene, serine/threonine kinase, related 206 |
| chr24_+_17270129 | 1.35 |
ENSDART00000186729
|
spag6
|
sperm associated antigen 6 |
| chr18_+_5543677 | 1.26 |
ENSDART00000146161
ENSDART00000136189 |
nnt2
|
nicotinamide nucleotide transhydrogenase 2 |
| chr7_-_1504382 | 1.25 |
ENSDART00000172770
|
si:zfos-405g10.4
|
si:zfos-405g10.4 |
| chr8_-_41273461 | 1.19 |
ENSDART00000190598
|
rnf10
|
ring finger protein 10 |
| chr25_-_32449235 | 1.17 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr7_+_23515966 | 1.17 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr12_-_25612170 | 1.17 |
ENSDART00000077155
|
six2b
|
SIX homeobox 2b |
| chr16_-_13613475 | 1.11 |
ENSDART00000139102
|
dbpb
|
D site albumin promoter binding protein b |
| chr14_+_33049579 | 1.10 |
ENSDART00000113470
|
tex11
|
testis expressed 11 |
| chr20_-_32658305 | 1.09 |
ENSDART00000140622
ENSDART00000152909 |
si:dkey-6f10.3
|
si:dkey-6f10.3 |
| chr22_+_36914636 | 1.09 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr7_-_24364536 | 1.09 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr14_+_3944826 | 1.09 |
ENSDART00000170167
|
lrp2bp
|
LRP2 binding protein |
| chr13_-_33683889 | 1.04 |
ENSDART00000136820
ENSDART00000065435 |
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr12_-_23129453 | 1.04 |
ENSDART00000077453
|
armc4
|
armadillo repeat containing 4 |
| chr12_+_26467847 | 1.02 |
ENSDART00000022495
|
ndel1a
|
nudE neurodevelopment protein 1-like 1a |
| chr8_-_13046089 | 0.98 |
ENSDART00000137784
|
si:dkey-208b23.5
|
si:dkey-208b23.5 |
| chr1_-_9109699 | 0.97 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr23_+_8797143 | 0.97 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr25_+_16646113 | 0.97 |
ENSDART00000110426
|
cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr8_-_21372446 | 0.96 |
ENSDART00000061481
ENSDART00000079293 |
ela2l
|
elastase 2 like |
| chr24_-_27419198 | 0.95 |
ENSDART00000141124
|
ccl34b.4
|
chemokine (C-C motif) ligand 34b, duplicate 4 |
| chr9_+_7732714 | 0.94 |
ENSDART00000145853
|
si:ch1073-349o24.2
|
si:ch1073-349o24.2 |
| chr2_+_24786765 | 0.94 |
ENSDART00000141030
|
pde4ca
|
phosphodiesterase 4C, cAMP-specific a |
| chr2_+_22694382 | 0.93 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr3_+_15809098 | 0.90 |
ENSDART00000183023
|
phospho1
|
phosphatase, orphan 1 |
| chr10_-_20453995 | 0.89 |
ENSDART00000168541
ENSDART00000164072 |
si:ch211-113d22.2
|
si:ch211-113d22.2 |
| chr13_-_6081803 | 0.88 |
ENSDART00000099224
|
dld
|
deltaD |
| chr5_-_21888368 | 0.88 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr7_-_20758825 | 0.88 |
ENSDART00000156717
ENSDART00000182629 ENSDART00000179801 |
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr17_-_10025234 | 0.87 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr7_-_16597130 | 0.87 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr16_+_31853919 | 0.86 |
ENSDART00000133886
|
atn1
|
atrophin 1 |
| chr12_-_23128746 | 0.83 |
ENSDART00000170018
|
armc4
|
armadillo repeat containing 4 |
| chr7_+_39706004 | 0.82 |
ENSDART00000161856
|
ccl36.1
|
chemokine (C-C motif) ligand 36, duplicate 1 |
| chr15_+_19838458 | 0.81 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr7_+_46368520 | 0.80 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr18_+_25752592 | 0.79 |
ENSDART00000111767
|
si:ch211-39k3.2
|
si:ch211-39k3.2 |
| chr3_+_26734162 | 0.79 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr24_-_33693070 | 0.77 |
ENSDART00000137483
|
si:dkeyp-87a6.2
|
si:dkeyp-87a6.2 |
| chr25_-_14637660 | 0.77 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr19_-_18152407 | 0.76 |
ENSDART00000193264
ENSDART00000016135 |
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr9_-_34885902 | 0.76 |
ENSDART00000137504
|
p2ry8
|
P2Y receptor family member 8 |
| chr1_+_50538839 | 0.76 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr17_-_33714636 | 0.76 |
ENSDART00000188500
|
DNAL1
|
si:dkey-84k17.3 |
| chr21_-_21790372 | 0.76 |
ENSDART00000151094
|
xrra1
|
X-ray radiation resistance associated 1 |
| chr20_+_33610271 | 0.75 |
ENSDART00000187186
|
CU467845.1
|
|
| chr11_-_22605981 | 0.75 |
ENSDART00000186923
|
myog
|
myogenin |
| chr12_-_36521767 | 0.75 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr19_+_19786117 | 0.75 |
ENSDART00000167757
ENSDART00000163546 |
hoxa1a
|
homeobox A1a |
| chr3_+_37197686 | 0.73 |
ENSDART00000151144
|
fmnl1a
|
formin-like 1a |
| chr14_-_21123551 | 0.73 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr23_-_29003864 | 0.73 |
ENSDART00000148257
|
casz1
|
castor zinc finger 1 |
| chr17_-_4395373 | 0.72 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr10_+_10351685 | 0.72 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr5_-_32445835 | 0.71 |
ENSDART00000170919
|
ncs1a
|
neuronal calcium sensor 1a |
| chr7_-_16596938 | 0.71 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr4_+_41602 | 0.70 |
ENSDART00000159640
|
phtf2
|
putative homeodomain transcription factor 2 |
| chr4_-_8152746 | 0.70 |
ENSDART00000012928
ENSDART00000177482 |
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr24_+_23730061 | 0.70 |
ENSDART00000080343
|
ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr25_+_25438165 | 0.69 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr10_+_31344227 | 0.69 |
ENSDART00000184470
|
AL845320.2
|
|
| chr2_+_31600661 | 0.68 |
ENSDART00000139039
|
si:ch211-106h4.4
|
si:ch211-106h4.4 |
| chr24_+_26658132 | 0.67 |
ENSDART00000183081
|
FQ378040.1
|
|
| chr5_+_28313824 | 0.66 |
ENSDART00000135878
|
si:dkeyp-86b9.1
|
si:dkeyp-86b9.1 |
| chr2_+_24722771 | 0.66 |
ENSDART00000147207
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr15_-_23376541 | 0.66 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr11_-_8208464 | 0.65 |
ENSDART00000161283
|
pimr203
|
Pim proto-oncogene, serine/threonine kinase, related 203 |
| chr16_-_54455573 | 0.65 |
ENSDART00000075275
|
pklr
|
pyruvate kinase L/R |
| chr7_-_51102479 | 0.64 |
ENSDART00000174023
|
col4a6
|
collagen, type IV, alpha 6 |
| chr20_+_33679500 | 0.64 |
ENSDART00000145242
|
si:ch211-199m3.1
|
si:ch211-199m3.1 |
| chr14_-_14566417 | 0.63 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr20_+_18225329 | 0.63 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr3_-_52674089 | 0.62 |
ENSDART00000154260
ENSDART00000125455 |
si:dkey-210j14.4
|
si:dkey-210j14.4 |
| chr14_-_21123325 | 0.62 |
ENSDART00000180889
ENSDART00000188539 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr20_+_33560419 | 0.61 |
ENSDART00000161563
ENSDART00000189650 |
si:dkey-65b13.8
|
si:dkey-65b13.8 |
| chr9_-_42696408 | 0.61 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr5_-_63425428 | 0.60 |
ENSDART00000163246
|
cntrl
|
centriolin |
| chr20_+_52458765 | 0.60 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr16_-_35585374 | 0.59 |
ENSDART00000180208
|
scmh1
|
Scm polycomb group protein homolog 1 |
| chr7_-_41403022 | 0.59 |
ENSDART00000174285
|
BX322787.1
|
|
| chr2_-_26563978 | 0.58 |
ENSDART00000150016
|
glis1a
|
GLIS family zinc finger 1a |
| chr15_+_37412883 | 0.58 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr4_-_9533345 | 0.58 |
ENSDART00000128501
|
si:ch73-139j3.4
|
si:ch73-139j3.4 |
| chr1_+_2101541 | 0.57 |
ENSDART00000128187
ENSDART00000167050 ENSDART00000182153 ENSDART00000122626 ENSDART00000164488 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr1_-_46505310 | 0.57 |
ENSDART00000178072
|
si:busm1-105l16.2
|
si:busm1-105l16.2 |
| chr8_-_41273768 | 0.56 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr4_-_26108053 | 0.56 |
ENSDART00000066951
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr2_-_16159491 | 0.56 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr4_-_8093753 | 0.56 |
ENSDART00000133434
|
wnk1b
|
WNK lysine deficient protein kinase 1b |
| chr20_-_1174266 | 0.56 |
ENSDART00000023304
|
gabrr2b
|
gamma-aminobutyric acid (GABA) A receptor, rho 2b |
| chr25_+_20119466 | 0.56 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr13_+_27316934 | 0.56 |
ENSDART00000164533
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr7_+_25858380 | 0.55 |
ENSDART00000148780
ENSDART00000079218 |
mtmr1a
|
myotubularin related protein 1a |
| chr22_-_30973791 | 0.55 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr9_-_46842179 | 0.55 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr10_+_2742499 | 0.53 |
ENSDART00000122847
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr21_+_25198637 | 0.52 |
ENSDART00000164972
|
si:dkey-183i3.6
|
si:dkey-183i3.6 |
| chr12_-_26851726 | 0.52 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr23_-_36724575 | 0.51 |
ENSDART00000159560
|
agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr14_-_8795081 | 0.51 |
ENSDART00000106671
|
tgfb2l
|
transforming growth factor, beta 2, like |
| chr22_-_36996856 | 0.51 |
ENSDART00000165923
|
ATP11B
|
si:dkey-211e20.10 |
| chr19_-_18152942 | 0.51 |
ENSDART00000190182
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr18_+_34861568 | 0.51 |
ENSDART00000192825
|
LO018333.1
|
|
| chr19_+_5604241 | 0.50 |
ENSDART00000011025
|
wipf2b
|
WAS/WASL interacting protein family, member 2b |
| chr11_+_24815667 | 0.50 |
ENSDART00000141730
|
rabif
|
RAB interacting factor |
| chr1_+_1689775 | 0.50 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 4 |
| chr11_+_13630107 | 0.50 |
ENSDART00000172220
|
si:ch211-1a19.3
|
si:ch211-1a19.3 |
| chr2_+_48288461 | 0.49 |
ENSDART00000141495
|
hes6
|
hes family bHLH transcription factor 6 |
| chr22_-_17729778 | 0.49 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr19_+_40856534 | 0.48 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr1_+_34203817 | 0.48 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr25_-_7925269 | 0.47 |
ENSDART00000014274
|
glcea
|
glucuronic acid epimerase a |
| chr20_+_35086242 | 0.47 |
ENSDART00000114262
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr7_+_19903924 | 0.47 |
ENSDART00000159112
|
si:ch211-285j22.3
|
si:ch211-285j22.3 |
| chr9_-_40765868 | 0.47 |
ENSDART00000138634
|
abca12
|
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr23_-_40796992 | 0.47 |
ENSDART00000145116
|
si:dkey-194e6.2
|
si:dkey-194e6.2 |
| chr4_+_11375894 | 0.47 |
ENSDART00000190471
ENSDART00000143963 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr7_-_40993456 | 0.47 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr10_-_36721726 | 0.46 |
ENSDART00000122132
|
pimr137
|
Pim proto-oncogene, serine/threonine kinase, related 137 |
| chr13_-_4333181 | 0.46 |
ENSDART00000122406
|
znf318
|
zinc finger protein 318 |
| chr25_-_7925019 | 0.46 |
ENSDART00000183309
|
glcea
|
glucuronic acid epimerase a |
| chr6_+_46431848 | 0.46 |
ENSDART00000181056
ENSDART00000144569 ENSDART00000064865 ENSDART00000133992 |
stau1
|
staufen double-stranded RNA binding protein 1 |
| chr16_+_50006145 | 0.46 |
ENSDART00000049375
|
ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr7_-_24995631 | 0.45 |
ENSDART00000173955
ENSDART00000173791 |
rcor2
|
REST corepressor 2 |
| chr20_+_35445462 | 0.45 |
ENSDART00000124497
|
tdrd6
|
tudor domain containing 6 |
| chr24_-_21923930 | 0.45 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr11_-_2478374 | 0.45 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr17_-_8899323 | 0.45 |
ENSDART00000081590
|
nkl.1
|
NK-lysin tandem duplicate 1 |
| chr25_-_3470910 | 0.44 |
ENSDART00000029067
ENSDART00000186737 |
hbp1
|
HMG-box transcription factor 1 |
| chr11_+_8129536 | 0.44 |
ENSDART00000158112
ENSDART00000011183 |
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr21_-_45073 | 0.44 |
ENSDART00000185997
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr9_+_38457806 | 0.43 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr4_-_52504961 | 0.43 |
ENSDART00000163123
|
si:dkey-78o7.3
|
si:dkey-78o7.3 |
| chr16_+_21330634 | 0.43 |
ENSDART00000191285
ENSDART00000183267 |
osbpl3b
|
oxysterol binding protein-like 3b |
| chr6_-_12314475 | 0.43 |
ENSDART00000156898
ENSDART00000157058 |
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr6_-_16394528 | 0.42 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr19_-_10656667 | 0.42 |
ENSDART00000081379
ENSDART00000151456 ENSDART00000143271 ENSDART00000182126 |
olah
|
oleoyl-ACP hydrolase |
| chr16_-_15263099 | 0.42 |
ENSDART00000125691
|
sntb1
|
syntrophin, basic 1 |
| chr21_+_11923701 | 0.42 |
ENSDART00000109292
|
ubap2a
|
ubiquitin associated protein 2a |
| chr2_+_14992879 | 0.42 |
ENSDART00000137546
|
pimr55
|
Pim proto-oncogene, serine/threonine kinase, related 55 |
| chr25_-_35996141 | 0.42 |
ENSDART00000149074
|
sall1b
|
spalt-like transcription factor 1b |
| chr5_+_62374092 | 0.42 |
ENSDART00000082965
|
CU928117.1
|
|
| chr19_+_25504645 | 0.41 |
ENSDART00000143292
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr19_-_7144548 | 0.41 |
ENSDART00000147177
ENSDART00000134850 |
psmb8a
psmb13a
|
proteasome subunit beta 8A proteasome subunit beta 13a |
| chr7_-_18656069 | 0.40 |
ENSDART00000021559
|
coro1b
|
coronin, actin binding protein, 1B |
| chr9_+_38458193 | 0.40 |
ENSDART00000008053
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr3_-_22212764 | 0.40 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr11_+_18216404 | 0.39 |
ENSDART00000086437
|
tmcc1b
|
transmembrane and coiled-coil domain family 1b |
| chr6_-_39764995 | 0.39 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr22_+_30839702 | 0.38 |
ENSDART00000139849
ENSDART00000135775 |
si:dkey-49n23.3
|
si:dkey-49n23.3 |
| chr2_-_32768951 | 0.38 |
ENSDART00000004712
|
bfsp2
|
beaded filament structural protein 2, phakinin |
| chr1_-_56213723 | 0.38 |
ENSDART00000142505
ENSDART00000137237 |
si:dkey-76b14.2
|
si:dkey-76b14.2 |
| chr5_+_65991152 | 0.38 |
ENSDART00000097756
|
lcn15
|
lipocalin 15 |
| chr8_-_17067364 | 0.38 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr17_+_23770848 | 0.38 |
ENSDART00000079646
|
kcnk18
|
potassium channel, subfamily K, member 18 |
| chr5_+_24059598 | 0.38 |
ENSDART00000051547
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr22_-_34937455 | 0.38 |
ENSDART00000169217
ENSDART00000188330 ENSDART00000165142 |
slit1b
|
slit homolog 1b (Drosophila) |
| chr17_-_23446760 | 0.38 |
ENSDART00000104715
|
pcgf5a
|
polycomb group ring finger 5a |
| chr18_+_31073265 | 0.38 |
ENSDART00000023539
|
cyba
|
cytochrome b-245, alpha polypeptide |
| chr1_-_47071979 | 0.38 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr3_+_31621774 | 0.37 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr21_+_33503835 | 0.37 |
ENSDART00000125658
|
clint1b
|
clathrin interactor 1b |
| chr2_+_24304854 | 0.37 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr6_+_45918981 | 0.37 |
ENSDART00000149642
|
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr6_+_9289802 | 0.37 |
ENSDART00000188650
|
kalrnb
|
kalirin RhoGEF kinase b |
| chr2_-_54054225 | 0.37 |
ENSDART00000167239
|
CABZ01050249.1
|
|
| chr7_+_65876335 | 0.37 |
ENSDART00000150143
|
tead1b
|
TEA domain family member 1b |
| chr4_+_20255160 | 0.37 |
ENSDART00000188658
|
lrtm2a
|
leucine-rich repeats and transmembrane domains 2a |
| chr8_+_21353878 | 0.36 |
ENSDART00000056420
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr5_-_39474235 | 0.36 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr2_+_40181633 | 0.36 |
ENSDART00000185494
|
FP236331.1
|
|
| chr5_+_26913120 | 0.36 |
ENSDART00000126609
|
tbx3b
|
T-box 3b |
| chr3_-_1204341 | 0.36 |
ENSDART00000089646
|
fam234b
|
family with sequence similarity 234, member B |
| chr4_-_15420452 | 0.36 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr1_+_10318089 | 0.36 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr1_-_10821584 | 0.35 |
ENSDART00000167452
ENSDART00000162137 |
crfb15
|
cytokine receptor family member B15 |
| chr16_-_22863348 | 0.35 |
ENSDART00000147609
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr4_-_26107841 | 0.35 |
ENSDART00000172012
|
si:ch211-244b2.4
|
si:ch211-244b2.4 |
| chr4_+_76735113 | 0.35 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr25_+_14109626 | 0.35 |
ENSDART00000109883
|
actc1c
|
actin, alpha, cardiac muscle 1c |
| chr3_+_60044780 | 0.35 |
ENSDART00000080437
|
zgc:113030
|
zgc:113030 |
| chr24_-_31843173 | 0.35 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr12_+_28856151 | 0.35 |
ENSDART00000152969
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr7_+_8324506 | 0.34 |
ENSDART00000168110
|
si:dkey-185m8.2
|
si:dkey-185m8.2 |
| chr10_-_5016997 | 0.34 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr19_+_1688727 | 0.34 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr7_-_33248773 | 0.34 |
ENSDART00000052389
|
itga11a
|
integrin, alpha 11a |
| chr4_+_77973876 | 0.34 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr17_+_6217704 | 0.34 |
ENSDART00000129100
|
BX000363.1
|
|
| chr2_-_16159203 | 0.34 |
ENSDART00000153480
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr23_-_16682186 | 0.34 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of runx2a+runx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.4 | 1.6 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.3 | 1.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.3 | 1.3 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.3 | 0.9 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.3 | 0.8 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.2 | 1.8 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 1.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.2 | 1.7 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 1.0 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.8 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 0.9 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.1 | 0.4 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.1 | 0.4 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.8 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.1 | 0.1 | GO:0031646 | positive regulation of neurological system process(GO:0031646) |
| 0.1 | 0.9 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.1 | 0.6 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.3 | GO:0036363 | transforming growth factor beta activation(GO:0036363) |
| 0.1 | 0.5 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.3 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.3 | GO:0019556 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 2.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.3 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 3.4 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.1 | 1.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 0.9 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 1.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.8 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 0.6 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.1 | 0.3 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.3 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.1 | 0.2 | GO:1900120 | regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.1 | 0.5 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.1 | 0.6 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.1 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.2 | GO:0003408 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) |
| 0.1 | 0.7 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.4 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.5 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.0 | 0.4 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0035093 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.8 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.3 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.4 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.2 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.4 | GO:0019320 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) hexose catabolic process(GO:0019320) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.3 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 1.8 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.3 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 2.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.2 | GO:0003151 | outflow tract morphogenesis(GO:0003151) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.4 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.4 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.0 | 0.3 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.0 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0046333 | octopamine biosynthetic process(GO:0006589) dopamine catabolic process(GO:0042420) norepinephrine biosynthetic process(GO:0042421) octopamine metabolic process(GO:0046333) |
| 0.0 | 0.7 | GO:0070167 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.1 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) retrograde trans-synaptic signaling by lipid(GO:0098920) retrograde trans-synaptic signaling by endocannabinoid(GO:0098921) |
| 0.0 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.8 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.2 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 1.0 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.4 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.1 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0071679 | commissural neuron axon guidance(GO:0071679) |
| 0.0 | 0.3 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.1 | GO:0002320 | lymphoid progenitor cell differentiation(GO:0002320) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.3 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.0 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.5 | GO:0032402 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.2 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.2 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.5 | GO:0050727 | regulation of inflammatory response(GO:0050727) |
| 0.0 | 3.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0099612 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.6 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.1 | GO:0000801 | central element(GO:0000801) |
| 0.2 | 0.8 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.2 | 1.8 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.1 | 1.8 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 0.6 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.1 | 1.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 1.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.2 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 0.4 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 0.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.5 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.8 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.0 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.3 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.4 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.5 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.6 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.6 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.2 | 0.9 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.2 | 1.7 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.6 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 0.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 1.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.1 | 0.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.6 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 0.4 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.3 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.1 | 1.3 | GO:0019870 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.1 | 0.2 | GO:0031701 | angiotensin receptor binding(GO:0031701) |
| 0.1 | 1.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 3.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 0.3 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.1 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.1 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0000810 | diacylglycerol diphosphate phosphatase activity(GO:0000810) |
| 0.0 | 1.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0043185 | vascular endothelial growth factor receptor 3 binding(GO:0043185) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.5 | GO:0005221 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0034596 | phosphatidylinositol phosphate 4-phosphatase activity(GO:0034596) |
| 0.0 | 0.2 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 1.1 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.3 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.3 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.5 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.1 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 2.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.4 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0019202 | amino acid kinase activity(GO:0019202) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.1 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.9 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 2.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.8 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.2 | 0.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.1 | 0.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.4 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.7 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.9 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.9 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 2.9 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.6 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 1.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |