Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
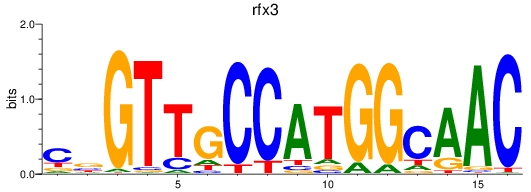
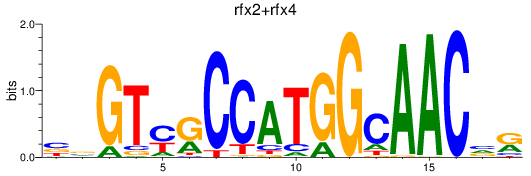
Results for rfx3_rfx2+rfx4
Z-value: 15.64
Transcription factors associated with rfx3_rfx2+rfx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx3
|
ENSDARG00000014550 | regulatory factor X, 3 (influences HLA class II expression) |
|
rfx2
|
ENSDARG00000013575 | regulatory factor X, 2 (influences HLA class II expression) |
|
rfx4
|
ENSDARG00000026395 | regulatory factor X, 4 |
|
rfx4
|
ENSDARG00000116861 | regulatory factor X, 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx3 | dr11_v1_chr10_-_641609_641609 | 0.99 | 1.2e-13 | Click! |
| rfx2 | dr11_v1_chr8_-_20138054_20138251 | 0.95 | 1.1e-09 | Click! |
| rfx4 | dr11_v1_chr18_-_15373620_15373620 | 0.17 | 5.0e-01 | Click! |
Activity profile of rfx3_rfx2+rfx4 motif
Sorted Z-values of rfx3_rfx2+rfx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_3915770 | 68.54 |
ENSDART00000159322
|
si:ch73-111k22.3
|
si:ch73-111k22.3 |
| chr3_-_1190132 | 66.60 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr4_-_5652030 | 59.33 |
ENSDART00000010903
|
rsph9
|
radial spoke head 9 homolog |
| chr4_-_13626831 | 58.00 |
ENSDART00000173077
|
si:dkeyp-81f3.4
|
si:dkeyp-81f3.4 |
| chr23_-_28692581 | 57.99 |
ENSDART00000078141
|
ribc1
|
RIB43A domain with coiled-coils 1 |
| chr25_+_21895182 | 55.95 |
ENSDART00000152075
|
si:ch211-147k9.8
|
si:ch211-147k9.8 |
| chr10_-_36682509 | 55.34 |
ENSDART00000148093
ENSDART00000063365 |
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr5_+_39563301 | 54.78 |
ENSDART00000131245
ENSDART00000097872 |
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr19_-_4023050 | 54.15 |
ENSDART00000162883
|
stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr16_+_4497302 | 53.67 |
ENSDART00000081826
ENSDART00000148096 |
ttc29
|
tetratricopeptide repeat domain 29 |
| chr25_-_10571078 | 53.41 |
ENSDART00000153898
|
si:ch211-107e6.5
|
si:ch211-107e6.5 |
| chr13_+_111212 | 53.08 |
ENSDART00000167840
|
dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr7_-_20778574 | 52.83 |
ENSDART00000078421
ENSDART00000190023 |
cyb5d1
|
cytochrome b5 domain containing 1 |
| chr5_-_57920600 | 52.78 |
ENSDART00000132376
|
si:dkey-27p23.3
|
si:dkey-27p23.3 |
| chr1_-_10806625 | 52.39 |
ENSDART00000139749
|
si:ch73-222h13.1
|
si:ch73-222h13.1 |
| chr20_+_33739059 | 51.69 |
ENSDART00000140361
|
ppil6
|
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr20_-_32658305 | 51.19 |
ENSDART00000140622
ENSDART00000152909 |
si:dkey-6f10.3
|
si:dkey-6f10.3 |
| chr20_-_13640598 | 50.29 |
ENSDART00000128823
ENSDART00000103394 |
rsph3
|
radial spoke 3 homolog |
| chr5_+_42386705 | 50.16 |
ENSDART00000143034
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr5_+_42407962 | 50.16 |
ENSDART00000188489
|
BX548073.11
|
|
| chr1_+_22427135 | 49.84 |
ENSDART00000147193
|
si:dkey-234d14.2
|
si:dkey-234d14.2 |
| chr5_-_42661012 | 49.46 |
ENSDART00000158339
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr3_+_31945604 | 49.40 |
ENSDART00000114957
|
zgc:193811
|
zgc:193811 |
| chr23_-_20133994 | 49.06 |
ENSDART00000004871
|
lrrc23
|
leucine rich repeat containing 23 |
| chr15_-_20939579 | 48.88 |
ENSDART00000152371
|
usp2a
|
ubiquitin specific peptidase 2a |
| chr17_-_39779906 | 48.61 |
ENSDART00000155181
|
pimr61
|
Pim proto-oncogene, serine/threonine kinase, related 61 |
| chr16_-_33930060 | 48.60 |
ENSDART00000110743
ENSDART00000101898 |
dnali1
|
dynein, axonemal, light intermediate chain 1 |
| chr5_+_42379517 | 48.38 |
ENSDART00000103325
|
pimr59
|
Pim proto-oncogene, serine/threonine kinase, related 59 |
| chr10_-_17232372 | 47.53 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr15_+_44053244 | 47.50 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr5_+_42393896 | 47.37 |
ENSDART00000189550
|
BX548073.13
|
|
| chr3_-_22829710 | 47.34 |
ENSDART00000055659
|
cyb561
|
cytochrome b561 |
| chr25_+_13157165 | 47.25 |
ENSDART00000155653
|
si:dkeyp-50b9.1
|
si:dkeyp-50b9.1 |
| chr5_+_42400777 | 47.10 |
ENSDART00000183114
|
BX548073.8
|
|
| chr12_-_145076 | 47.02 |
ENSDART00000160926
|
dnah9
|
dynein, axonemal, heavy chain 9 |
| chr21_-_22557469 | 46.08 |
ENSDART00000167230
|
cfap53
|
cilia and flagella associated protein 53 |
| chr3_+_60716904 | 46.02 |
ENSDART00000168280
|
foxj1a
|
forkhead box J1a |
| chr5_-_23200880 | 45.90 |
ENSDART00000051531
|
iqcd
|
IQ motif containing D |
| chr17_-_39772999 | 45.77 |
ENSDART00000155727
|
pimr60
|
Pim proto-oncogene, serine/threonine kinase, related 60 |
| chr10_-_17231917 | 45.49 |
ENSDART00000038577
|
rab36
|
RAB36, member RAS oncogene family |
| chr2_-_14272083 | 45.18 |
ENSDART00000169986
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr6_-_21582444 | 45.08 |
ENSDART00000151339
|
si:dkey-43k4.3
|
si:dkey-43k4.3 |
| chr14_+_3944826 | 44.71 |
ENSDART00000170167
|
lrp2bp
|
LRP2 binding protein |
| chr13_+_9408101 | 44.16 |
ENSDART00000091502
|
si:ch211-243o19.4
|
si:ch211-243o19.4 |
| chr9_+_9441453 | 43.96 |
ENSDART00000081859
ENSDART00000188567 ENSDART00000143101 |
maats1
|
MYCBP-associated, testis expressed 1 |
| chr8_+_23809843 | 43.79 |
ENSDART00000099724
|
si:ch211-163l21.10
|
si:ch211-163l21.10 |
| chr16_-_45069882 | 43.35 |
ENSDART00000058384
|
gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr24_+_13277573 | 43.08 |
ENSDART00000137886
|
si:ch211-171b20.3
|
si:ch211-171b20.3 |
| chr2_-_1569250 | 42.22 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr25_+_37480285 | 42.16 |
ENSDART00000166187
|
CABZ01095001.1
|
|
| chr11_-_12437234 | 41.95 |
ENSDART00000169324
|
zgc:174355
|
zgc:174355 |
| chr21_+_18292535 | 41.48 |
ENSDART00000170205
ENSDART00000169676 ENSDART00000163063 |
dnai1.1
|
dynein, axonemal, intermediate chain 1, paralog 1 |
| chr7_+_41295974 | 41.44 |
ENSDART00000173568
ENSDART00000173544 |
si:dkey-86l18.10
|
si:dkey-86l18.10 |
| chr5_-_71705191 | 41.35 |
ENSDART00000187767
|
ak1
|
adenylate kinase 1 |
| chr13_+_33117528 | 40.28 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr21_+_17956856 | 40.17 |
ENSDART00000080431
|
dnai1.2
|
dynein, axonemal, intermediate chain 1, paralog 2 |
| chr13_-_36703164 | 39.11 |
ENSDART00000044357
|
cdkl1
|
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
| chr25_-_28638052 | 39.09 |
ENSDART00000138918
ENSDART00000135247 ENSDART00000114662 ENSDART00000157493 ENSDART00000137677 |
agbl2
|
ATP/GTP binding protein-like 2 |
| chr13_+_33304187 | 38.47 |
ENSDART00000075826
ENSDART00000145295 |
dcdc2b
|
doublecortin domain containing 2B |
| chr17_-_24684687 | 38.07 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr8_-_44586981 | 37.92 |
ENSDART00000026831
ENSDART00000113945 |
rsph14
|
radial spoke head 14 homolog |
| chr11_-_12379541 | 37.86 |
ENSDART00000171717
|
zgc:174353
|
zgc:174353 |
| chr13_+_24717880 | 37.80 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr11_-_12717004 | 37.61 |
ENSDART00000122020
|
si:ch211-209f22.3
|
si:ch211-209f22.3 |
| chr5_-_8406192 | 37.29 |
ENSDART00000159718
|
spef2
|
sperm flagellar 2 |
| chr25_+_13731542 | 37.27 |
ENSDART00000161012
|
ccdc135
|
coiled-coil domain containing 135 |
| chr6_+_120181 | 37.25 |
ENSDART00000151209
ENSDART00000185930 |
cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr2_-_14271874 | 37.13 |
ENSDART00000189390
|
tctex1d1
|
Tctex1 domain containing 1 |
| chr1_+_57269441 | 37.08 |
ENSDART00000149688
|
mycbpap
|
mycbp associated protein |
| chr20_-_2667902 | 36.60 |
ENSDART00000036373
|
cfap206
|
cilia and flagella associated protein 206 |
| chr21_+_22423286 | 36.27 |
ENSDART00000133190
|
capslb
|
calcyphosine-like b |
| chr11_-_12334546 | 36.27 |
ENSDART00000127768
|
zgc:91850
|
zgc:91850 |
| chr4_-_390431 | 36.10 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr2_+_35728033 | 35.88 |
ENSDART00000002094
|
ankrd45
|
ankyrin repeat domain 45 |
| chr13_-_6218248 | 35.70 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr21_+_22828500 | 35.42 |
ENSDART00000151109
|
si:rp71-1p14.7
|
si:rp71-1p14.7 |
| chr17_-_45164291 | 35.40 |
ENSDART00000062109
|
noxred1
|
NADP-dependent oxidoreductase domain containing 1 |
| chr10_-_29744921 | 35.15 |
ENSDART00000088605
|
ift46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr5_-_54672763 | 34.79 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr10_-_9410068 | 34.08 |
ENSDART00000041382
|
morn5
|
MORN repeat containing 5 |
| chr11_+_11430552 | 33.68 |
ENSDART00000160339
|
si:dkey-23f9.11
|
si:dkey-23f9.11 |
| chr2_-_33455164 | 33.38 |
ENSDART00000134024
ENSDART00000132221 |
ccdc24
|
coiled-coil domain containing 24 |
| chr5_+_56965177 | 33.19 |
ENSDART00000097409
ENSDART00000157239 |
tmem232
|
transmembrane protein 232 |
| chr11_-_12364122 | 32.94 |
ENSDART00000170129
|
zgc:174353
|
zgc:174353 |
| chr25_+_32525131 | 32.93 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr17_+_14425219 | 32.92 |
ENSDART00000153994
|
ccdc175
|
coiled-coil domain containing 175 |
| chr5_+_66353750 | 32.85 |
ENSDART00000143410
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr2_+_2181709 | 32.74 |
ENSDART00000092763
ENSDART00000148553 |
ccdc13
|
coiled-coil domain containing 13 |
| chr18_-_41232297 | 32.66 |
ENSDART00000036928
|
fbxo36a
|
F-box protein 36a |
| chr19_-_40186328 | 32.32 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr20_+_27691307 | 32.17 |
ENSDART00000153299
|
si:dkey-1h6.8
|
si:dkey-1h6.8 |
| chr17_-_8753354 | 32.09 |
ENSDART00000154593
|
tex36
|
testis expressed 36 |
| chr12_-_47793857 | 31.51 |
ENSDART00000161294
|
dydc2
|
DPY30 domain containing 2 |
| chr16_-_54640315 | 31.17 |
ENSDART00000101207
|
spag1b
|
sperm associated antigen 1b |
| chr23_+_35708730 | 31.09 |
ENSDART00000009277
|
tuba1a
|
tubulin, alpha 1a |
| chr8_-_13315304 | 31.07 |
ENSDART00000142596
|
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr11_-_12499116 | 31.03 |
ENSDART00000134836
|
si:dkey-27d5.4
|
si:dkey-27d5.4 |
| chr22_-_25033105 | 31.00 |
ENSDART00000124220
|
nptxrb
|
neuronal pentraxin receptor b |
| chr7_-_28403930 | 30.84 |
ENSDART00000076496
|
stk33
|
serine/threonine kinase 33 |
| chr1_+_11711226 | 30.73 |
ENSDART00000147623
|
si:dkey-26i13.8
|
si:dkey-26i13.8 |
| chr5_-_65969959 | 30.68 |
ENSDART00000170677
|
ttc16
|
tetratricopeptide repeat domain 16 |
| chr15_+_33865312 | 30.12 |
ENSDART00000166141
|
tekt1
|
tektin 1 |
| chr1_-_17715493 | 30.07 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr11_-_12483302 | 29.97 |
ENSDART00000135031
|
si:dkey-27d5.5
|
si:dkey-27d5.5 |
| chr11_-_12350419 | 29.94 |
ENSDART00000166732
|
si:dkey-27d5.14
|
si:dkey-27d5.14 |
| chr6_+_26309968 | 29.80 |
ENSDART00000153805
|
dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr24_-_5911973 | 29.56 |
ENSDART00000077933
ENSDART00000077922 |
pimr64
|
Pim proto-oncogene, serine/threonine kinase, related 64 |
| chr22_+_2937485 | 29.49 |
ENSDART00000082222
ENSDART00000135507 ENSDART00000143258 |
cep19
|
centrosomal protein 19 |
| chr11_-_12394682 | 29.44 |
ENSDART00000172447
|
si:dkey-27d5.10
|
si:dkey-27d5.10 |
| chr7_-_41295356 | 29.25 |
ENSDART00000074175
|
si:dkey-86l18.8
|
si:dkey-86l18.8 |
| chr18_-_7097403 | 29.18 |
ENSDART00000003748
|
cfap161
|
cilia and flagella associated protein 161 |
| chr8_-_13315567 | 29.13 |
ENSDART00000132685
ENSDART00000168635 |
kcnn1b
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1b |
| chr7_+_21321317 | 28.80 |
ENSDART00000173950
|
dnah2
|
dynein, axonemal, heavy chain 2 |
| chr18_+_30028135 | 28.74 |
ENSDART00000140825
ENSDART00000145306 ENSDART00000136810 |
si:ch211-220f16.1
|
si:ch211-220f16.1 |
| chr11_-_12363941 | 28.71 |
ENSDART00000192304
|
zgc:174353
|
zgc:174353 |
| chr7_-_52388734 | 28.66 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr8_-_7504146 | 28.63 |
ENSDART00000148339
ENSDART00000140098 |
cdk20
|
cyclin-dependent kinase 20 |
| chr6_-_16804001 | 28.60 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr18_+_46382484 | 28.55 |
ENSDART00000024202
ENSDART00000142790 |
daw1
|
dynein assembly factor with WDR repeat domains 1 |
| chr5_+_66353589 | 28.52 |
ENSDART00000138246
|
si:ch211-261c8.5
|
si:ch211-261c8.5 |
| chr5_-_33022014 | 28.38 |
ENSDART00000061149
|
zgc:55461
|
zgc:55461 |
| chr20_+_18225329 | 28.32 |
ENSDART00000144172
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr24_+_26345427 | 28.27 |
ENSDART00000089756
|
lrrc34
|
leucine rich repeat containing 34 |
| chr9_-_27771339 | 28.24 |
ENSDART00000135722
ENSDART00000140381 |
si:rp71-45g20.11
|
si:rp71-45g20.11 |
| chr17_-_6600899 | 28.22 |
ENSDART00000154074
ENSDART00000180912 |
ANKRD66
|
si:ch211-189e2.2 |
| chr11_-_12530304 | 28.13 |
ENSDART00000143061
|
zgc:174354
|
zgc:174354 |
| chr2_+_19578079 | 27.97 |
ENSDART00000144413
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr2_+_19633493 | 27.93 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr23_+_31245395 | 27.84 |
ENSDART00000053588
|
irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr14_+_23518110 | 27.76 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr11_-_12379359 | 27.65 |
ENSDART00000187551
|
zgc:174353
|
zgc:174353 |
| chr3_-_12026741 | 27.64 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr9_-_44289636 | 27.52 |
ENSDART00000110411
|
cerkl
|
ceramide kinase-like |
| chr18_-_27897217 | 27.36 |
ENSDART00000175259
|
iqcg
|
IQ motif containing G |
| chr7_-_41295511 | 27.28 |
ENSDART00000173455
|
si:dkey-86l18.8
|
si:dkey-86l18.8 |
| chr24_+_17269849 | 27.23 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr6_+_15762647 | 27.09 |
ENSDART00000127133
ENSDART00000128939 |
iqca1
|
IQ motif containing with AAA domain 1 |
| chr6_+_7250824 | 27.04 |
ENSDART00000177226
|
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr11_-_12619789 | 26.88 |
ENSDART00000185508
|
CR450764.10
|
|
| chr8_+_18615938 | 26.76 |
ENSDART00000089161
|
si:ch211-232d19.4
|
si:ch211-232d19.4 |
| chr2_+_19522082 | 26.63 |
ENSDART00000146098
|
pimr49
|
Pim proto-oncogene, serine/threonine kinase, related 49 |
| chr23_+_12134839 | 26.39 |
ENSDART00000128551
ENSDART00000141204 |
ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr11_-_12472006 | 26.32 |
ENSDART00000160558
|
si:dkey-27d5.6
|
si:dkey-27d5.6 |
| chr10_-_34889053 | 26.10 |
ENSDART00000136966
|
ccdc169
|
coiled-coil domain containing 169 |
| chr25_+_13731726 | 25.97 |
ENSDART00000181065
|
ccdc135
|
coiled-coil domain containing 135 |
| chr18_+_21273749 | 25.91 |
ENSDART00000143265
|
hydin
|
HYDIN, axonemal central pair apparatus protein |
| chr4_+_20177526 | 25.90 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr9_-_16853462 | 25.75 |
ENSDART00000160273
|
CT573248.2
|
|
| chr4_-_76154252 | 25.43 |
ENSDART00000161269
|
BX957252.1
|
|
| chr17_+_7513673 | 25.39 |
ENSDART00000156674
|
klhl10b.1
|
kelch-like family member 10b, tandem duplicate 1 |
| chr17_+_46864116 | 25.28 |
ENSDART00000156250
|
pimr27
|
Pim proto-oncogene, serine/threonine kinase, related 27 |
| chr21_-_22828593 | 24.95 |
ENSDART00000150993
|
angptl5
|
angiopoietin-like 5 |
| chr14_+_8725216 | 24.67 |
ENSDART00000157630
|
pimr57
|
Pim proto-oncogene, serine/threonine kinase, related 57 |
| chr6_+_46258866 | 24.67 |
ENSDART00000134584
|
zgc:162324
|
zgc:162324 |
| chr2_+_52122663 | 24.61 |
ENSDART00000111266
|
theg
|
theg spermatid protein |
| chr2_-_19630297 | 24.61 |
ENSDART00000100111
ENSDART00000142173 |
pimr51
BX088713.2
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr25_+_27873671 | 24.60 |
ENSDART00000088817
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr24_-_6678640 | 24.45 |
ENSDART00000042478
|
enkur
|
enkurin, TRPC channel interacting protein |
| chr8_+_21195420 | 24.44 |
ENSDART00000100234
ENSDART00000091307 |
col2a1a
|
collagen, type II, alpha 1a |
| chr2_-_19576640 | 24.33 |
ENSDART00000141021
|
pimr51
|
Pim proto-oncogene, serine/threonine kinase, related 51 |
| chr11_-_12437052 | 24.32 |
ENSDART00000190890
ENSDART00000183980 |
zgc:174355
|
zgc:174355 |
| chr19_+_46240171 | 24.25 |
ENSDART00000162785
|
mapk15
|
mitogen-activated protein kinase 15 |
| chr11_-_12634017 | 24.23 |
ENSDART00000158286
ENSDART00000193090 |
CR450764.1
|
|
| chr5_+_23190642 | 24.21 |
ENSDART00000144846
|
cfap73
|
cilia and flagella associated protein 73 |
| chr1_+_49651016 | 24.18 |
ENSDART00000074380
ENSDART00000101017 |
tsga10
|
testis specific, 10 |
| chr9_+_23748342 | 23.84 |
ENSDART00000019053
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr18_+_35173683 | 23.75 |
ENSDART00000192545
|
cfap45
|
cilia and flagella associated protein 45 |
| chr24_+_5912635 | 23.73 |
ENSDART00000153736
|
pimr63
|
Pim proto-oncogene, serine/threonine kinase, related 63 |
| chr17_+_28882977 | 23.72 |
ENSDART00000153937
|
prkd1
|
protein kinase D1 |
| chr23_+_13928346 | 23.57 |
ENSDART00000155326
|
si:dkey-90a13.10
|
si:dkey-90a13.10 |
| chr17_-_46817295 | 23.52 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr13_+_9408445 | 23.47 |
ENSDART00000162415
|
si:ch211-243o19.4
|
si:ch211-243o19.4 |
| chr8_+_53204027 | 23.42 |
ENSDART00000135353
|
CFAP74
|
cilia and flagella associated protein 74 |
| chr15_-_18162647 | 23.30 |
ENSDART00000012064
|
pih1d2
|
PIH1 domain containing 2 |
| chr25_+_27873836 | 23.18 |
ENSDART00000163801
|
iqub
|
IQ motif and ubiquitin domain containing |
| chr10_-_29236860 | 23.18 |
ENSDART00000111620
|
ccdc83
|
coiled-coil domain containing 83 |
| chr9_+_23748133 | 23.17 |
ENSDART00000180758
|
faima
|
Fas apoptotic inhibitory molecule a |
| chr18_+_35173859 | 23.14 |
ENSDART00000127379
ENSDART00000098292 |
cfap45
|
cilia and flagella associated protein 45 |
| chr6_+_16895685 | 23.03 |
ENSDART00000154090
|
pimr28
|
Pim proto-oncogene, serine/threonine kinase, related 28 |
| chr11_-_12471837 | 22.98 |
ENSDART00000113175
ENSDART00000186965 |
si:dkey-27d5.6
|
si:dkey-27d5.6 |
| chr19_-_9472893 | 22.75 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr1_-_3295045 | 22.55 |
ENSDART00000157572
|
gpc5a
|
glypican 5a |
| chr11_-_12301939 | 22.43 |
ENSDART00000184239
|
CABZ01022564.1
|
|
| chr14_-_36799280 | 22.35 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr13_-_13662624 | 22.25 |
ENSDART00000143269
|
pimr47
|
Pim proto-oncogene, serine/threonine kinase, related 47 |
| chr18_-_29962234 | 22.24 |
ENSDART00000144996
|
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr20_+_591505 | 22.18 |
ENSDART00000046438
|
kcnk2b
|
potassium channel, subfamily K, member 2b |
| chr6_+_16870004 | 22.17 |
ENSDART00000154794
|
pimr21
|
Pim proto-oncogene, serine/threonine kinase, related 21 |
| chr11_-_12394500 | 22.15 |
ENSDART00000140221
|
si:dkey-27d5.10
|
si:dkey-27d5.10 |
| chr5_+_22633188 | 22.11 |
ENSDART00000128781
|
mtnr1c
|
melatonin receptor 1C |
| chr14_+_30762131 | 22.07 |
ENSDART00000145039
|
si:ch211-145o7.3
|
si:ch211-145o7.3 |
| chr2_-_29761965 | 21.95 |
ENSDART00000142057
ENSDART00000110595 |
si:dkey-188g12.1
|
si:dkey-188g12.1 |
| chr20_-_7080427 | 21.95 |
ENSDART00000140166
ENSDART00000023677 |
si:ch211-121a2.2
|
si:ch211-121a2.2 |
| chr18_-_29961784 | 21.93 |
ENSDART00000135601
ENSDART00000140661 |
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr10_+_31222656 | 21.87 |
ENSDART00000140988
ENSDART00000143387 |
tmem218
|
transmembrane protein 218 |
| chr17_+_53250802 | 21.75 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr2_+_19578446 | 21.70 |
ENSDART00000164758
|
pimr50
|
Pim proto-oncogene, serine/threonine kinase, related 50 |
| chr17_-_46933567 | 21.61 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr20_+_20637866 | 21.59 |
ENSDART00000060203
ENSDART00000079079 |
rtn1b
|
reticulon 1b |
| chr24_+_26345609 | 21.48 |
ENSDART00000186844
|
lrrc34
|
leucine rich repeat containing 34 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx3_rfx2+rfx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 22.2 | 155.5 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 19.8 | 59.3 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 12.5 | 62.7 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 9.8 | 39.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 9.2 | 27.5 | GO:2000425 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) regulation of apoptotic cell clearance(GO:2000425) |
| 8.2 | 238.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 8.1 | 57.0 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 7.6 | 15.2 | GO:0090660 | cerebrospinal fluid circulation(GO:0090660) |
| 7.0 | 49.1 | GO:0032475 | otolith formation(GO:0032475) |
| 6.5 | 39.2 | GO:0089700 | protein kinase D signaling(GO:0089700) regulation of lymphangiogenesis(GO:1901490) |
| 5.9 | 41.4 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 5.9 | 29.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 5.7 | 68.5 | GO:0001881 | receptor recycling(GO:0001881) |
| 5.4 | 54.2 | GO:1902686 | positive regulation of mitochondrial membrane permeability(GO:0035794) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 5.3 | 42.2 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 5.0 | 95.3 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 4.5 | 45.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 4.5 | 63.2 | GO:0030317 | sperm motility(GO:0030317) |
| 4.4 | 57.3 | GO:0035082 | axoneme assembly(GO:0035082) |
| 4.3 | 30.1 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 4.3 | 47.0 | GO:1902042 | regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902041) negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 4.2 | 54.7 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 3.9 | 35.4 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 3.9 | 66.6 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 3.5 | 31.5 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 3.5 | 145.0 | GO:0003341 | cilium movement(GO:0003341) |
| 3.0 | 11.8 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 2.7 | 8.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 2.6 | 929.8 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 2.5 | 15.3 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 2.5 | 27.7 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 2.4 | 4.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 2.2 | 26.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 2.0 | 15.6 | GO:0036372 | opsin transport(GO:0036372) |
| 1.9 | 5.7 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 1.8 | 38.5 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 1.8 | 7.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 1.7 | 55.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 1.7 | 51.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 1.7 | 36.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 1.6 | 11.4 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 1.6 | 17.7 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 1.6 | 12.8 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 1.5 | 22.7 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 1.3 | 24.0 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 1.3 | 749.6 | GO:0006397 | mRNA processing(GO:0006397) |
| 1.3 | 5.2 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 1.3 | 16.7 | GO:0030819 | activation of adenylate cyclase activity(GO:0007190) positive regulation of cAMP metabolic process(GO:0030816) positive regulation of cAMP biosynthetic process(GO:0030819) positive regulation of adenylate cyclase activity(GO:0045762) |
| 1.2 | 7.4 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.2 | 10.7 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 1.1 | 27.8 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 1.1 | 16.5 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 1.0 | 16.3 | GO:0036065 | fucosylation(GO:0036065) |
| 1.0 | 14.1 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 1.0 | 21.1 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 1.0 | 16.4 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) mitotic DNA integrity checkpoint(GO:0044774) |
| 1.0 | 77.4 | GO:0006165 | nucleoside diphosphate phosphorylation(GO:0006165) |
| 0.9 | 3.8 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.9 | 3.8 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.9 | 7.5 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.9 | 4.7 | GO:0070378 | ERK5 cascade(GO:0070375) regulation of ERK5 cascade(GO:0070376) positive regulation of ERK5 cascade(GO:0070378) |
| 0.9 | 68.4 | GO:0044782 | cilium organization(GO:0044782) |
| 0.8 | 8.1 | GO:0071941 | nitrogen cycle metabolic process(GO:0071941) |
| 0.8 | 28.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.7 | 9.4 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.7 | 18.8 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.7 | 16.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.7 | 27.0 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.7 | 4.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.6 | 3.6 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.6 | 28.4 | GO:1902749 | regulation of cell cycle G2/M phase transition(GO:1902749) |
| 0.6 | 21.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.6 | 20.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.5 | 6.0 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.5 | 15.1 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.5 | 17.3 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.5 | 13.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.5 | 5.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.5 | 9.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.5 | 22.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.5 | 60.6 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.5 | 20.7 | GO:0051057 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.5 | 3.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.4 | 8.9 | GO:0006415 | translational termination(GO:0006415) |
| 0.4 | 8.2 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.4 | 1.7 | GO:0030718 | RNA 5'-end processing(GO:0000966) germ-line stem cell population maintenance(GO:0030718) |
| 0.4 | 86.0 | GO:0060271 | cilium morphogenesis(GO:0060271) |
| 0.4 | 12.5 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.4 | 4.0 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.4 | 4.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.4 | 1.8 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.4 | 11.8 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.3 | 17.4 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.3 | 13.9 | GO:1902850 | mitotic spindle assembly(GO:0090307) microtubule cytoskeleton organization involved in mitosis(GO:1902850) |
| 0.3 | 18.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.3 | 17.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.3 | 15.4 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.3 | 10.6 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.3 | 0.9 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.3 | 22.0 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.3 | 11.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.3 | 2.3 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.3 | 3.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.3 | 41.6 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 2.7 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.2 | 16.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.2 | 9.3 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.2 | 14.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.2 | 11.0 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.2 | 14.3 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.2 | 3.7 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.2 | 0.6 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 | 25.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.2 | 2.0 | GO:0015886 | heme transport(GO:0015886) iron coordination entity transport(GO:1901678) |
| 0.2 | 0.8 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.2 | 22.9 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.2 | 1.7 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 12.4 | GO:0007249 | I-kappaB kinase/NF-kappaB signaling(GO:0007249) |
| 0.2 | 19.2 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.2 | 3.2 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.1 | 1.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 6.6 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.1 | 11.0 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 6.7 | GO:0051781 | positive regulation of cell division(GO:0051781) |
| 0.1 | 1.1 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 6.0 | GO:0001764 | neuron migration(GO:0001764) |
| 0.1 | 6.0 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.1 | 1.9 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.1 | 6.2 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 8.2 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 10.1 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 1.5 | GO:0015780 | nucleotide-sugar transport(GO:0015780) pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 7.3 | GO:0030042 | actin filament depolymerization(GO:0030042) |
| 0.1 | 2.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 11.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 12.4 | GO:0043066 | negative regulation of apoptotic process(GO:0043066) |
| 0.1 | 0.2 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 6.4 | GO:0032886 | regulation of microtubule-based process(GO:0032886) regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.1 | 1.2 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.1 | 0.7 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 5.0 | GO:0030865 | cortical cytoskeleton organization(GO:0030865) |
| 0.1 | 2.7 | GO:0008593 | regulation of Notch signaling pathway(GO:0008593) |
| 0.1 | 2.4 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.1 | 4.0 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.1 | 0.7 | GO:0035094 | response to nicotine(GO:0035094) response to alkaloid(GO:0043279) |
| 0.1 | 8.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.0 | 0.8 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 2.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.2 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.1 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 1.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 1.5 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.3 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 2.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 7.7 | GO:0000278 | mitotic cell cycle(GO:0000278) |
| 0.0 | 11.7 | GO:0006886 | intracellular protein transport(GO:0006886) |
| 0.0 | 2.7 | GO:0009116 | nucleoside metabolic process(GO:0009116) |
| 0.0 | 0.3 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 0.2 | GO:0014846 | vascular endothelial growth factor production(GO:0010573) regulation of vascular endothelial growth factor production(GO:0010574) positive regulation of vascular endothelial growth factor production(GO:0010575) esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 32.0 | 95.9 | GO:0001534 | radial spoke(GO:0001534) |
| 13.5 | 54.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 8.5 | 102.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 6.4 | 58.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 5.8 | 23.3 | GO:0097255 | R2TP complex(GO:0097255) |
| 5.6 | 66.6 | GO:1990246 | uniplex complex(GO:1990246) |
| 5.2 | 382.2 | GO:0031514 | motile cilium(GO:0031514) |
| 4.9 | 34.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 4.6 | 50.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 4.2 | 163.3 | GO:0005930 | axoneme(GO:0005930) |
| 4.1 | 32.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 3.5 | 21.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 3.5 | 24.5 | GO:0097223 | sperm part(GO:0097223) |
| 2.8 | 13.9 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 2.6 | 162.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 2.3 | 27.6 | GO:0036038 | MKS complex(GO:0036038) |
| 1.8 | 31.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 1.7 | 11.8 | GO:1990130 | Iml1 complex(GO:1990130) |
| 1.6 | 11.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 1.5 | 6.0 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 1.4 | 20.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 1.3 | 15.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 1.2 | 18.6 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 1.1 | 213.7 | GO:0005929 | cilium(GO:0005929) |
| 1.1 | 12.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.9 | 22.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.8 | 38.5 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.8 | 13.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.7 | 11.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.6 | 4.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.6 | 27.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.5 | 3.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.5 | 68.5 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.5 | 57.9 | GO:0030658 | transport vesicle membrane(GO:0030658) |
| 0.5 | 46.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.5 | 12.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.4 | 20.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.4 | 13.3 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.4 | 8.4 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.3 | 1.7 | GO:1990923 | PET complex(GO:1990923) |
| 0.3 | 2.3 | GO:0030892 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 3.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.3 | 38.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.3 | 16.3 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 5.3 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 6.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 78.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.2 | 22.1 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 8.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 13.7 | GO:0001726 | ruffle(GO:0001726) |
| 0.2 | 2.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 1.6 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 9.0 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.1 | 15.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 13.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.1 | 5.1 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 93.6 | GO:0005829 | cytosol(GO:0005829) |
| 0.1 | 4.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 536.6 | GO:0005737 | cytoplasm(GO:0005737) |
| 0.1 | 9.3 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 4.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 537.4 | GO:0005634 | nucleus(GO:0005634) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 9.0 | GO:0045202 | synapse(GO:0045202) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.8 | 47.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 10.8 | 43.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 7.1 | 35.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 6.9 | 130.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 6.5 | 58.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 5.7 | 74.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 5.3 | 21.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 4.3 | 12.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 3.9 | 27.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 3.7 | 22.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 3.2 | 58.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 2.7 | 67.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 2.6 | 33.3 | GO:0070840 | dynein complex binding(GO:0070840) |
| 2.5 | 80.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 1.9 | 15.5 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.9 | 807.2 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 1.9 | 26.4 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 1.6 | 12.8 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 1.5 | 56.5 | GO:0051087 | chaperone binding(GO:0051087) |
| 1.5 | 22.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 1.5 | 39.2 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 1.4 | 39.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.4 | 14.2 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 1.4 | 16.7 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 1.3 | 43.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 1.2 | 7.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 1.2 | 71.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 1.2 | 8.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 1.1 | 12.6 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 1.1 | 19.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 1.1 | 20.2 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 1.0 | 934.6 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 1.0 | 14.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.0 | 49.3 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 1.0 | 16.3 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 1.0 | 4.8 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.9 | 8.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.9 | 9.4 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.9 | 4.7 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) |
| 0.9 | 8.9 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.9 | 35.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.7 | 13.8 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.7 | 11.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.7 | 17.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.7 | 8.2 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.7 | 17.7 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 7.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.6 | 13.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 6.8 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.5 | 15.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.5 | 13.6 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.5 | 27.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.5 | 67.3 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.5 | 3.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.5 | 21.2 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.5 | 4.0 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.5 | 6.3 | GO:0005222 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.5 | 57.4 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.5 | 3.2 | GO:0016880 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.4 | 7.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 20.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.4 | 5.7 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.4 | 3.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.4 | 32.3 | GO:0008080 | N-acetyltransferase activity(GO:0008080) |
| 0.4 | 1.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 54.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.4 | 28.7 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.3 | 31.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 135.6 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.3 | 1.6 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.3 | 41.9 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.3 | 5.6 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.3 | 26.0 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.3 | 4.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.3 | 2.7 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.3 | 3.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 5.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.0 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 39.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 10.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.2 | 10.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 0.9 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.2 | 29.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 34.6 | GO:0016820 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.2 | 1.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.2 | 4.4 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 3.8 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 3.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 1.6 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.7 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.2 | 3.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 1.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 7.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 19.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 32.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.1 | 25.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 5.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 6.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 1.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.2 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 19.6 | GO:0016887 | ATPase activity(GO:0016887) |
| 0.1 | 6.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 10.2 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.1 | 1.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 28.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 3.7 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 1.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.9 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.5 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.5 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 36.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 1.2 | 21.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 1.1 | 39.2 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 1.0 | 36.0 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.9 | 52.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.8 | 16.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.7 | 7.5 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.5 | 6.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.5 | 15.2 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.2 | 8.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.2 | 26.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.1 | 2.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.6 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.1 | 10.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 2.1 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 1.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 32.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 2.5 | 15.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 2.1 | 43.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 2.0 | 41.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.9 | 31.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 1.9 | 16.7 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 1.4 | 39.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 1.2 | 12.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 1.0 | 34.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.8 | 13.6 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.5 | 4.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.4 | 6.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.4 | 5.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.4 | 4.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.3 | 16.4 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 11.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 11.0 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.2 | 5.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 6.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 6.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 9.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.1 | 2.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |