Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
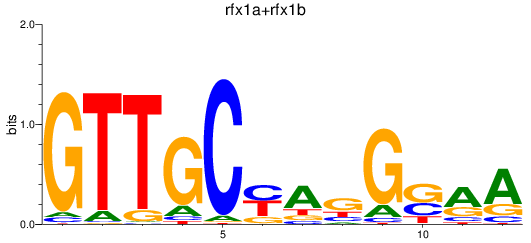
Results for rfx1a+rfx1b
Z-value: 2.17
Transcription factors associated with rfx1a+rfx1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx1a
|
ENSDARG00000005883 | regulatory factor X, 1a (influences HLA class II expression) |
|
rfx1b
|
ENSDARG00000075904 | regulatory factor X, 1b (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx1a | dr11_v1_chr3_-_19200571_19200651 | 0.99 | 1.2e-14 | Click! |
| rfx1b | dr11_v1_chr1_-_54107321_54107321 | 0.94 | 4.4e-09 | Click! |
Activity profile of rfx1a+rfx1b motif
Sorted Z-values of rfx1a+rfx1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_48219111 | 10.17 |
ENSDART00000111225
ENSDART00000145972 |
ccdc173
|
coiled-coil domain containing 173 |
| chr5_+_61919217 | 9.46 |
ENSDART00000097340
|
rsph4a
|
radial spoke head 4 homolog A |
| chr6_-_8264751 | 9.20 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr4_+_15819728 | 8.80 |
ENSDART00000101613
|
lrguk
|
leucine-rich repeats and guanylate kinase domain containing |
| chr25_+_21895182 | 8.46 |
ENSDART00000152075
|
si:ch211-147k9.8
|
si:ch211-147k9.8 |
| chr1_+_22002649 | 8.40 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr5_-_29671586 | 8.01 |
ENSDART00000098336
|
spaca9
|
sperm acrosome associated 9 |
| chr14_+_6954579 | 7.96 |
ENSDART00000060998
|
nme5
|
NME/NM23 family member 5 |
| chr5_+_39563301 | 7.94 |
ENSDART00000131245
ENSDART00000097872 |
si:ch211-57h10.1
|
si:ch211-57h10.1 |
| chr1_-_6225285 | 7.90 |
ENSDART00000141653
|
mdh1b
|
malate dehydrogenase 1B, NAD (soluble) |
| chr20_-_3915770 | 7.82 |
ENSDART00000159322
|
si:ch73-111k22.3
|
si:ch73-111k22.3 |
| chr20_-_31075972 | 7.23 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr22_+_3223489 | 7.00 |
ENSDART00000082011
|
lim2.2
|
lens intrinsic membrane protein 2.2 |
| chr20_+_21022641 | 6.82 |
ENSDART00000152561
ENSDART00000141591 |
si:dkey-219e20.2
|
si:dkey-219e20.2 |
| chr1_+_47831619 | 6.77 |
ENSDART00000110335
ENSDART00000101066 |
cfap58
|
cilia and flagella associated protein 58 |
| chr5_+_29671681 | 6.62 |
ENSDART00000043096
|
ak8
|
adenylate kinase 8 |
| chr10_-_29744921 | 6.42 |
ENSDART00000088605
|
ift46
|
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr17_+_27545183 | 6.22 |
ENSDART00000129392
|
pacrg
|
PARK2 co-regulated |
| chr2_-_58085444 | 6.08 |
ENSDART00000166395
|
fcer1gl
|
Fc receptor, IgE, high affinity I, gamma polypeptide like |
| chr17_-_33716688 | 6.03 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr12_-_145076 | 5.96 |
ENSDART00000160926
|
dnah9
|
dynein, axonemal, heavy chain 9 |
| chr11_-_12232929 | 5.83 |
ENSDART00000127611
|
si:ch211-156l18.4
|
si:ch211-156l18.4 |
| chr2_-_1569250 | 5.75 |
ENSDART00000167202
|
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr10_-_17232372 | 5.46 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr8_+_53204027 | 5.42 |
ENSDART00000135353
|
CFAP74
|
cilia and flagella associated protein 74 |
| chr11_-_12549110 | 5.38 |
ENSDART00000148014
|
zgc:174354
|
zgc:174354 |
| chr15_+_22267847 | 5.35 |
ENSDART00000110665
|
spa17
|
sperm autoantigenic protein 17 |
| chr24_-_37003577 | 5.35 |
ENSDART00000137777
|
dnaaf3
|
dynein, axonemal, assembly factor 3 |
| chr18_+_2168695 | 5.35 |
ENSDART00000165855
|
dnaaf4
|
dynein axonemal assembly factor 4 |
| chr13_+_24717880 | 5.30 |
ENSDART00000147713
|
cfap43
|
cilia and flagella associated protein 43 |
| chr7_+_23515966 | 5.24 |
ENSDART00000186893
ENSDART00000186189 |
zgc:109889
|
zgc:109889 |
| chr10_-_36682509 | 5.21 |
ENSDART00000148093
ENSDART00000063365 |
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr17_+_11500628 | 5.11 |
ENSDART00000155660
|
efcab2
|
EF-hand calcium binding domain 2 |
| chr6_+_515181 | 5.10 |
ENSDART00000171374
|
CACNA1I (1 of many)
|
si:ch73-379f7.5 |
| chr20_-_4766645 | 5.06 |
ENSDART00000147071
ENSDART00000152398 |
ak7a
|
adenylate kinase 7a |
| chr3_-_60175470 | 5.03 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
| chr7_-_34505985 | 4.99 |
ENSDART00000173806
ENSDART00000024869 |
tex9
|
testis expressed 9 |
| chr7_-_40738774 | 4.97 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr3_-_61593274 | 4.97 |
ENSDART00000154132
ENSDART00000055071 |
nptx2a
|
neuronal pentraxin 2a |
| chr2_+_7075220 | 4.91 |
ENSDART00000022963
|
cdc14aa
|
cell division cycle 14Aa |
| chr5_+_32260502 | 4.88 |
ENSDART00000149020
|
si:ch211-158m24.12
|
si:ch211-158m24.12 |
| chr1_+_50538839 | 4.87 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr15_-_23206562 | 4.85 |
ENSDART00000110360
|
ccdc153
|
coiled-coil domain containing 153 |
| chr19_+_9091673 | 4.83 |
ENSDART00000052898
|
si:ch211-81a5.5
|
si:ch211-81a5.5 |
| chr13_+_33117528 | 4.82 |
ENSDART00000085719
|
si:ch211-10a23.2
|
si:ch211-10a23.2 |
| chr19_+_27589201 | 4.77 |
ENSDART00000182060
|
si:dkeyp-46h3.1
|
si:dkeyp-46h3.1 |
| chr25_-_4192637 | 4.62 |
ENSDART00000153832
|
ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr10_-_17231917 | 4.62 |
ENSDART00000038577
|
rab36
|
RAB36, member RAS oncogene family |
| chr24_-_17067284 | 4.54 |
ENSDART00000111237
|
armc3
|
armadillo repeat containing 3 |
| chr15_+_44053244 | 4.51 |
ENSDART00000059550
|
lrrc51
|
leucine rich repeat containing 51 |
| chr17_-_4395373 | 4.49 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr25_-_28638052 | 4.45 |
ENSDART00000138918
ENSDART00000135247 ENSDART00000114662 ENSDART00000157493 ENSDART00000137677 |
agbl2
|
ATP/GTP binding protein-like 2 |
| chr11_-_30352333 | 4.33 |
ENSDART00000030794
|
tmem169a
|
transmembrane protein 169a |
| chr4_-_12795436 | 4.32 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr2_+_13462305 | 4.30 |
ENSDART00000149309
ENSDART00000080900 |
cfap57
|
cilia and flagella associated protein 57 |
| chr1_+_22003173 | 4.27 |
ENSDART00000054388
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr2_-_7696503 | 4.25 |
ENSDART00000169709
|
CABZ01055306.1
|
|
| chr17_+_23937262 | 4.23 |
ENSDART00000113276
|
si:ch211-189k9.2
|
si:ch211-189k9.2 |
| chr13_+_48482256 | 4.21 |
ENSDART00000053332
|
FOXN2 (1 of many)
|
forkhead box N2 |
| chr7_+_38445938 | 4.20 |
ENSDART00000173467
|
cep89
|
centrosomal protein 89 |
| chr11_+_37652870 | 4.18 |
ENSDART00000129918
|
kif17
|
kinesin family member 17 |
| chr22_+_25049563 | 4.08 |
ENSDART00000078173
|
dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr11_-_12417952 | 4.06 |
ENSDART00000135694
|
si:dkey-27d5.9
|
si:dkey-27d5.9 |
| chr19_-_40186328 | 4.01 |
ENSDART00000087474
|
efcab1
|
EF-hand calcium binding domain 1 |
| chr7_-_28403930 | 4.00 |
ENSDART00000076496
|
stk33
|
serine/threonine kinase 33 |
| chr8_-_50525360 | 3.99 |
ENSDART00000175648
|
CABZ01060030.1
|
|
| chr11_-_12512122 | 3.97 |
ENSDART00000145338
|
si:dkey-27d5.3
|
si:dkey-27d5.3 |
| chr20_-_32007209 | 3.96 |
ENSDART00000021575
|
adgb
|
androglobin |
| chr12_-_36521767 | 3.83 |
ENSDART00000110290
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr6_+_18251140 | 3.81 |
ENSDART00000169752
|
ccdc40
|
coiled-coil domain containing 40 |
| chr24_+_2843268 | 3.77 |
ENSDART00000170529
|
si:ch211-152c8.2
|
si:ch211-152c8.2 |
| chr2_-_7696287 | 3.77 |
ENSDART00000190769
|
CABZ01055306.1
|
|
| chr19_+_17400283 | 3.76 |
ENSDART00000127353
|
nr1d2b
|
nuclear receptor subfamily 1, group D, member 2b |
| chr1_+_11711226 | 3.68 |
ENSDART00000147623
|
si:dkey-26i13.8
|
si:dkey-26i13.8 |
| chr12_-_11649690 | 3.57 |
ENSDART00000149713
|
btbd16
|
BTB (POZ) domain containing 16 |
| chr17_-_24684687 | 3.49 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr13_-_6218248 | 3.46 |
ENSDART00000159052
|
si:zfos-1056e6.1
|
si:zfos-1056e6.1 |
| chr5_+_43807003 | 3.45 |
ENSDART00000097625
|
zgc:158640
|
zgc:158640 |
| chr13_+_24679674 | 3.43 |
ENSDART00000033090
ENSDART00000139854 |
zgc:66426
|
zgc:66426 |
| chr12_+_17620067 | 3.20 |
ENSDART00000073599
|
rsph10b
|
radial spoke head 10 homolog B |
| chr23_-_42752550 | 3.19 |
ENSDART00000187059
|
si:ch73-217n20.1
|
si:ch73-217n20.1 |
| chr6_-_40842768 | 3.19 |
ENSDART00000076160
|
mustn1a
|
musculoskeletal, embryonic nuclear protein 1a |
| chr18_-_31051847 | 3.19 |
ENSDART00000170982
|
gas8
|
growth arrest-specific 8 |
| chr17_+_53250802 | 3.13 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr10_-_7988396 | 3.06 |
ENSDART00000141445
ENSDART00000024282 |
ewsr1a
|
EWS RNA-binding protein 1a |
| chr18_+_16192083 | 3.01 |
ENSDART00000133042
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr4_+_20177526 | 2.97 |
ENSDART00000017947
ENSDART00000135451 |
ccdc146
|
coiled-coil domain containing 146 |
| chr20_-_12642685 | 2.95 |
ENSDART00000173257
|
si:dkey-97l20.6
|
si:dkey-97l20.6 |
| chr2_-_42960353 | 2.89 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr14_+_24283915 | 2.86 |
ENSDART00000172868
|
klhl3
|
kelch-like family member 3 |
| chr11_+_11374021 | 2.82 |
ENSDART00000170806
|
si:dkey-23f9.8
|
si:dkey-23f9.8 |
| chr14_+_35892802 | 2.79 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr25_+_25438165 | 2.79 |
ENSDART00000161369
|
LRRC56
|
si:ch211-103e16.5 |
| chr15_-_1022436 | 2.76 |
ENSDART00000156003
|
znf1010
|
zinc finger protein 1010 |
| chr13_-_7767548 | 2.70 |
ENSDART00000158720
ENSDART00000121952 |
h2afy2
|
H2A histone family, member Y2 |
| chr13_-_48764180 | 2.69 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr3_-_12026741 | 2.63 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr10_-_10864331 | 2.62 |
ENSDART00000122657
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr17_-_31695217 | 2.57 |
ENSDART00000104332
ENSDART00000143090 |
lin52
|
lin-52 DREAM MuvB core complex component |
| chr25_+_18436301 | 2.53 |
ENSDART00000056180
|
cep41
|
centrosomal protein 41 |
| chr5_+_37087583 | 2.51 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr3_-_57425961 | 2.51 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr11_+_11347031 | 2.51 |
ENSDART00000172429
ENSDART00000193128 |
si:dkey-23f9.6
|
si:dkey-23f9.6 |
| chr3_+_24458899 | 2.50 |
ENSDART00000156655
|
cbx6b
|
chromobox homolog 6b |
| chr13_-_31470439 | 2.49 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr10_-_10863936 | 2.47 |
ENSDART00000180568
|
nrarpa
|
NOTCH regulated ankyrin repeat protein a |
| chr16_-_54640315 | 2.44 |
ENSDART00000101207
|
spag1b
|
sperm associated antigen 1b |
| chr8_+_7144066 | 2.34 |
ENSDART00000146306
|
slc6a6a
|
solute carrier family 6 (neurotransmitter transporter), member 6a |
| chr24_-_36910224 | 2.28 |
ENSDART00000079233
|
dnaaf3l
|
dynein, axonemal, assembly factor 3 like |
| chr16_+_34493987 | 2.26 |
ENSDART00000138374
|
si:ch211-255i3.4
|
si:ch211-255i3.4 |
| chr23_+_21638258 | 2.23 |
ENSDART00000104188
|
igsf21b
|
immunoglobin superfamily, member 21b |
| chr2_-_29761965 | 2.23 |
ENSDART00000142057
ENSDART00000110595 |
si:dkey-188g12.1
|
si:dkey-188g12.1 |
| chr1_+_8662530 | 2.22 |
ENSDART00000054989
|
fscn1b
|
fascin actin-bundling protein 1b |
| chr8_-_50147948 | 2.22 |
ENSDART00000149010
|
hp
|
haptoglobin |
| chr11_+_11360641 | 2.21 |
ENSDART00000159765
|
si:dkey-23f9.7
|
si:dkey-23f9.7 |
| chr9_-_40873934 | 2.19 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr25_+_35942867 | 2.17 |
ENSDART00000066985
|
hsd17b2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr20_+_30797329 | 2.16 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr11_-_12232769 | 2.09 |
ENSDART00000180116
|
si:ch211-156l18.4
|
si:ch211-156l18.4 |
| chr19_-_10648523 | 2.08 |
ENSDART00000059358
|
tekt2
|
tektin 2 (testicular) |
| chr14_+_24277556 | 2.07 |
ENSDART00000122660
|
hnrnpa0a
|
heterogeneous nuclear ribonucleoprotein A0a |
| chr3_-_6719232 | 2.06 |
ENSDART00000154294
|
atg4db
|
autophagy related 4D, cysteine peptidase b |
| chr3_+_60044780 | 2.04 |
ENSDART00000080437
|
zgc:113030
|
zgc:113030 |
| chr5_-_33022014 | 2.03 |
ENSDART00000061149
|
zgc:55461
|
zgc:55461 |
| chr1_+_54683655 | 2.03 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr25_+_25438322 | 1.97 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr17_-_29119362 | 1.96 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr8_-_36469117 | 1.95 |
ENSDART00000111240
|
mhc2dab
|
major histocompatibility complex class II DAB gene |
| chr6_+_52350443 | 1.93 |
ENSDART00000151612
ENSDART00000151349 |
si:ch211-239j9.1
|
si:ch211-239j9.1 |
| chr17_+_33767890 | 1.93 |
ENSDART00000193177
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr10_+_2529037 | 1.93 |
ENSDART00000123467
|
zmp:0000001301
|
zmp:0000001301 |
| chr1_-_45633955 | 1.92 |
ENSDART00000044057
|
sept3
|
septin 3 |
| chr6_-_7711349 | 1.91 |
ENSDART00000032494
|
ttc26
|
tetratricopeptide repeat domain 26 |
| chr7_-_50395059 | 1.88 |
ENSDART00000191150
|
vps33b
|
vacuolar protein sorting 33B |
| chr11_+_44723019 | 1.86 |
ENSDART00000168531
|
irf2bp2b
|
interferon regulatory factor 2 binding protein 2b |
| chr24_+_39186940 | 1.85 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr2_-_43123949 | 1.81 |
ENSDART00000075347
ENSDART00000132346 |
lrrc6
|
leucine rich repeat containing 6 |
| chr9_+_9112159 | 1.81 |
ENSDART00000165932
|
sik1
|
salt-inducible kinase 1 |
| chr4_-_2545310 | 1.76 |
ENSDART00000150619
ENSDART00000140760 |
e2f7
|
E2F transcription factor 7 |
| chr9_+_40874194 | 1.74 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr12_-_36521947 | 1.70 |
ENSDART00000152946
|
unc13d
|
unc-13 homolog D (C. elegans) |
| chr11_-_18799827 | 1.70 |
ENSDART00000185438
ENSDART00000189116 ENSDART00000180504 |
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr24_-_37472727 | 1.68 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr13_-_47556447 | 1.62 |
ENSDART00000131239
|
acoxl
|
acyl-CoA oxidase-like |
| chr20_-_46554440 | 1.59 |
ENSDART00000043298
ENSDART00000060680 |
fosab
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Ab |
| chr6_+_13933464 | 1.58 |
ENSDART00000109144
|
ptprnb
|
protein tyrosine phosphatase, receptor type, Nb |
| chr8_+_25267903 | 1.57 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr22_-_10891213 | 1.57 |
ENSDART00000145229
|
arhgef18b
|
rho/rac guanine nucleotide exchange factor (GEF) 18b |
| chr1_-_49498116 | 1.55 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr24_-_39772045 | 1.53 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr2_+_6126638 | 1.50 |
ENSDART00000153916
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr23_+_44157682 | 1.50 |
ENSDART00000164474
ENSDART00000149928 |
si:ch73-106g13.1
|
si:ch73-106g13.1 |
| chr14_+_1351214 | 1.50 |
ENSDART00000007091
|
bbs12
|
Bardet-Biedl syndrome 12 |
| chr5_-_52813442 | 1.50 |
ENSDART00000169305
|
zgc:158260
|
zgc:158260 |
| chr18_-_17531483 | 1.48 |
ENSDART00000061000
|
bbs2
|
Bardet-Biedl syndrome 2 |
| chr20_+_4060839 | 1.47 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr24_+_40860320 | 1.43 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr21_-_22317920 | 1.39 |
ENSDART00000191083
ENSDART00000108701 |
gdpd4b
|
glycerophosphodiester phosphodiesterase domain containing 4b |
| chr1_+_11711503 | 1.36 |
ENSDART00000138355
|
si:dkey-26i13.8
|
si:dkey-26i13.8 |
| chr4_-_78026285 | 1.36 |
ENSDART00000168273
|
CCT2
|
chaperonin containing TCP1 subunit 2 |
| chr17_+_21964472 | 1.32 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr15_+_13984879 | 1.27 |
ENSDART00000159438
|
zgc:162730
|
zgc:162730 |
| chr16_+_11779534 | 1.26 |
ENSDART00000133497
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr7_+_27059330 | 1.26 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr5_+_69856153 | 1.25 |
ENSDART00000124128
ENSDART00000073663 |
ugt2a6
|
UDP glucuronosyltransferase 2 family, polypeptide A6 |
| chr1_-_21538673 | 1.24 |
ENSDART00000131257
|
wdr19
|
WD repeat domain 19 |
| chr4_+_77971104 | 1.24 |
ENSDART00000188609
|
zgc:113921
|
zgc:113921 |
| chr16_+_11779761 | 1.24 |
ENSDART00000140297
|
si:dkey-250k15.4
|
si:dkey-250k15.4 |
| chr4_+_13615731 | 1.22 |
ENSDART00000141395
ENSDART00000144310 |
ccdc87
|
coiled-coil domain containing 87 |
| chr14_-_21219659 | 1.20 |
ENSDART00000089867
|
ppp2r2cb
|
protein phosphatase 2, regulatory subunit B, gamma b |
| chr17_+_43595692 | 1.16 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr11_-_18800299 | 1.15 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr23_+_2760573 | 1.15 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr13_+_28854438 | 1.12 |
ENSDART00000193407
ENSDART00000189554 |
CU639469.1
|
|
| chr21_+_21612214 | 1.11 |
ENSDART00000008099
|
b9d2
|
B9 domain containing 2 |
| chr5_-_52243424 | 1.06 |
ENSDART00000159078
|
erap2
|
endoplasmic reticulum aminopeptidase 2 |
| chr18_-_43884044 | 1.06 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr1_+_55662491 | 1.02 |
ENSDART00000152386
|
adgre8
|
adhesion G protein-coupled receptor E8 |
| chr1_-_21538329 | 1.00 |
ENSDART00000182798
ENSDART00000193518 ENSDART00000183873 ENSDART00000183376 |
wdr19
|
WD repeat domain 19 |
| chr8_-_49345388 | 0.99 |
ENSDART00000053203
|
plp2
|
proteolipid protein 2 |
| chr21_+_9576176 | 0.97 |
ENSDART00000161289
ENSDART00000159899 ENSDART00000162834 |
mapk10
|
mitogen-activated protein kinase 10 |
| chr21_+_21611867 | 0.97 |
ENSDART00000189148
|
b9d2
|
B9 domain containing 2 |
| chr10_-_44482911 | 0.97 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr8_-_19216180 | 0.95 |
ENSDART00000146162
|
si:ch73-222f22.2
|
si:ch73-222f22.2 |
| chr16_+_46111849 | 0.95 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr4_-_23858900 | 0.94 |
ENSDART00000123199
|
usp6nl
|
USP6 N-terminal like |
| chr12_+_4971515 | 0.92 |
ENSDART00000161076
|
arhgap27
|
Rho GTPase activating protein 27 |
| chr16_+_9726038 | 0.91 |
ENSDART00000192988
ENSDART00000020859 |
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr15_+_37589698 | 0.91 |
ENSDART00000076066
ENSDART00000153894 ENSDART00000156298 |
lin37
|
lin-37 DREAM MuvB core complex component |
| chr10_+_25204626 | 0.90 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr17_-_10838434 | 0.88 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr24_-_9960290 | 0.88 |
ENSDART00000143390
ENSDART00000092975 ENSDART00000184953 |
vps41
|
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr18_+_29950233 | 0.86 |
ENSDART00000146431
|
atmin
|
ATM interactor |
| chr11_-_24428237 | 0.85 |
ENSDART00000189107
ENSDART00000103752 |
dvl1b
|
dishevelled segment polarity protein 1b |
| chr19_-_8604429 | 0.84 |
ENSDART00000151165
|
trim46b
|
tripartite motif containing 46b |
| chr24_+_744713 | 0.84 |
ENSDART00000067764
|
stk17a
|
serine/threonine kinase 17a |
| chr4_-_9557186 | 0.84 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr16_+_5202042 | 0.82 |
ENSDART00000145368
|
soga3a
|
SOGA family member 3a |
| chr11_+_1575435 | 0.81 |
ENSDART00000155713
ENSDART00000156562 |
si:dkey-40c23.1
|
si:dkey-40c23.1 |
| chr10_+_37981471 | 0.75 |
ENSDART00000113112
ENSDART00000185102 |
wscd1b
|
WSC domain containing 1b |
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx1a+rfx1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:1902176 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 1.8 | 5.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 1.6 | 4.9 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 1.6 | 7.9 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 1.1 | 4.4 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 1.1 | 22.5 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 1.0 | 11.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 1.0 | 2.9 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.8 | 3.1 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.8 | 5.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.7 | 5.7 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.7 | 7.8 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.6 | 16.1 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.6 | 3.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.6 | 1.9 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.6 | 4.2 | GO:0098971 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.5 | 3.2 | GO:0035889 | otolith tethering(GO:0035889) |
| 0.5 | 4.5 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.4 | 14.0 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.4 | 5.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.4 | 1.6 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 2.3 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.4 | 2.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.4 | 1.8 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.3 | 2.1 | GO:0036372 | opsin transport(GO:0036372) |
| 0.3 | 2.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.3 | 4.3 | GO:0045824 | negative regulation of toll-like receptor signaling pathway(GO:0034122) negative regulation of innate immune response(GO:0045824) |
| 0.3 | 1.7 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.3 | 2.7 | GO:0000022 | mitotic spindle elongation(GO:0000022) positive regulation of cytokinesis(GO:0032467) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.3 | 1.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.2 | 2.0 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 10.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.2 | 3.5 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 0.6 | GO:0090278 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.2 | 2.9 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.2 | 2.5 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 2.2 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 2.9 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.2 | 0.9 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) positive regulation of calcium ion import(GO:0090280) |
| 0.2 | 5.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 | 11.7 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
| 0.1 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.1 | 1.5 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 1.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.9 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 0.9 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.1 | 1.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 | 1.6 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 1.6 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 4.0 | GO:0042770 | signal transduction in response to DNA damage(GO:0042770) |
| 0.1 | 1.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.6 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.2 | GO:1901232 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 1.9 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.1 | 0.5 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 1.1 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.7 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 5.6 | GO:0044782 | cilium organization(GO:0044782) |
| 0.1 | 2.3 | GO:0072348 | sulfur compound transport(GO:0072348) |
| 0.1 | 2.4 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 1.1 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.0 | 1.8 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 3.1 | GO:0051966 | regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 | 0.3 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.2 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 1.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 19.4 | GO:0006397 | mRNA processing(GO:0006397) |
| 0.0 | 1.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.6 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.3 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.0 | 2.2 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.1 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.7 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 1.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.0 | GO:0006986 | response to unfolded protein(GO:0006986) |
| 0.0 | 0.5 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 1.0 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 2.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.6 | GO:0046890 | regulation of lipid biosynthetic process(GO:0046890) |
| 0.0 | 1.1 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 9.5 | GO:0001534 | radial spoke(GO:0001534) |
| 2.0 | 6.1 | GO:0032998 | Fc receptor complex(GO:0032997) Fc-epsilon receptor I complex(GO:0032998) |
| 0.8 | 8.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.8 | 14.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.8 | 4.5 | GO:0060091 | kinocilium(GO:0060091) kinociliary basal body(GO:1902636) |
| 0.7 | 4.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 2.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.4 | 1.9 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.4 | 5.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.3 | 12.9 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.3 | 17.1 | GO:0031514 | motile cilium(GO:0031514) |
| 0.3 | 18.5 | GO:0030286 | dynein complex(GO:0030286) |
| 0.3 | 4.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 0.7 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 2.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 0.9 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 1.1 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 7.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 0.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.9 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 3.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 1.9 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.1 | 5.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.1 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.6 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 1.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 15.1 | GO:0005829 | cytosol(GO:0005829) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.3 | 11.7 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 2.0 | 6.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) IgE receptor activity(GO:0019767) |
| 1.8 | 8.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 1.6 | 7.9 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.9 | 5.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.7 | 5.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 2.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.5 | 6.0 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.4 | 6.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.4 | 12.4 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.4 | 2.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.3 | 8.0 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.3 | 4.9 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.3 | 1.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.3 | 1.5 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.3 | 9.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.2 | 4.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.2 | 1.8 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 2.9 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.2 | 2.5 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 0.9 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.2 | 4.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 5.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.6 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 4.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 1.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 1.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 6.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.7 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.1 | 28.1 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.3 | GO:1901682 | sulfur compound transmembrane transporter activity(GO:1901682) |
| 0.1 | 4.6 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 4.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.6 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.0 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.2 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.0 | 0.5 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 5.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 3.8 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 10.9 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 1.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 6.3 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 1.3 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.5 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 1.7 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.1 | 1.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 1.8 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.9 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 1.0 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.1 | 1.7 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.1 | 1.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.6 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |