Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
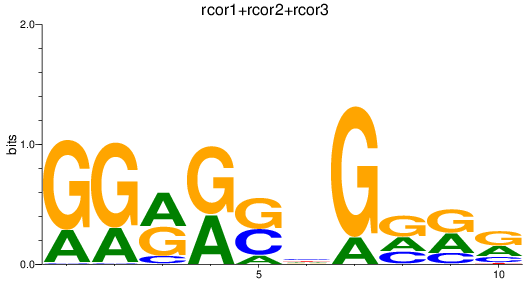
Results for rcor1+rcor2+rcor3
Z-value: 0.32
Transcription factors associated with rcor1+rcor2+rcor3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rcor3
|
ENSDARG00000004502 | REST corepressor 3 |
|
rcor2
|
ENSDARG00000008278 | REST corepressor 2 |
|
rcor1
|
ENSDARG00000031434 | REST corepressor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rcor2 | dr11_v1_chr7_-_24995631_24995640 | 0.57 | 1.4e-02 | Click! |
| rcor1 | dr11_v1_chr17_-_29271359_29271359 | 0.09 | 7.1e-01 | Click! |
| rcor3 | dr11_v1_chr22_+_38276024_38276024 | -0.01 | 9.8e-01 | Click! |
Activity profile of rcor1+rcor2+rcor3 motif
Sorted Z-values of rcor1+rcor2+rcor3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_13822137 | 0.33 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr15_+_31806303 | 0.25 |
ENSDART00000155902
|
frya
|
furry homolog a (Drosophila) |
| chr11_-_6206520 | 0.23 |
ENSDART00000150199
ENSDART00000148246 ENSDART00000019440 |
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr17_+_26783828 | 0.22 |
ENSDART00000154921
|
larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr12_-_33659328 | 0.21 |
ENSDART00000153457
|
tmem94
|
transmembrane protein 94 |
| chr19_-_2029777 | 0.19 |
ENSDART00000128639
|
CABZ01071939.1
|
|
| chr22_-_24984733 | 0.17 |
ENSDART00000142147
ENSDART00000187284 |
dnal4b
|
dynein, axonemal, light chain 4b |
| chr20_-_32981053 | 0.17 |
ENSDART00000138708
|
nbas
|
neuroblastoma amplified sequence |
| chr6_-_60104628 | 0.17 |
ENSDART00000057463
ENSDART00000169188 |
pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_+_1467863 | 0.16 |
ENSDART00000173433
|
emc4
|
ER membrane protein complex subunit 4 |
| chr25_-_37258653 | 0.16 |
ENSDART00000131076
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr22_+_12366516 | 0.16 |
ENSDART00000157802
|
r3hdm1
|
R3H domain containing 1 |
| chr8_+_13106760 | 0.15 |
ENSDART00000029308
|
itgb4
|
integrin, beta 4 |
| chr3_+_60957512 | 0.15 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr20_-_46808870 | 0.14 |
ENSDART00000190911
|
esrrgb
|
estrogen-related receptor gamma b |
| chr16_+_11992498 | 0.14 |
ENSDART00000143442
|
p3h3
|
prolyl 3-hydroxylase 3 |
| chr18_+_19772874 | 0.13 |
ENSDART00000043455
|
smad3b
|
SMAD family member 3b |
| chr21_-_41369370 | 0.13 |
ENSDART00000159290
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr12_-_4475890 | 0.13 |
ENSDART00000092492
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr11_+_807153 | 0.13 |
ENSDART00000173289
|
vgll4b
|
vestigial-like family member 4b |
| chr19_+_6938289 | 0.13 |
ENSDART00000139122
ENSDART00000178832 |
flot1b
|
flotillin 1b |
| chr21_-_43398457 | 0.13 |
ENSDART00000166530
|
ccni2
|
cyclin I family, member 2 |
| chr6_+_41255485 | 0.12 |
ENSDART00000042683
ENSDART00000186013 |
cadpsb
|
Ca2+-dependent activator protein for secretion b |
| chr1_-_53700392 | 0.12 |
ENSDART00000127216
|
fam161a
|
family with sequence similarity 161, member A |
| chr5_-_15692030 | 0.11 |
ENSDART00000099545
|
CR384085.1
|
|
| chr8_-_42712573 | 0.11 |
ENSDART00000132229
ENSDART00000137258 |
si:ch73-138n13.1
|
si:ch73-138n13.1 |
| chr1_+_14454663 | 0.11 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr3_+_60007703 | 0.11 |
ENSDART00000157351
ENSDART00000153928 ENSDART00000155876 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr23_-_6722101 | 0.11 |
ENSDART00000157828
|
baz2a
|
bromodomain adjacent to zinc finger domain, 2A |
| chr12_-_4408828 | 0.11 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr22_-_38508112 | 0.11 |
ENSDART00000192004
|
klc4
|
kinesin light chain 4 |
| chr7_-_13906409 | 0.11 |
ENSDART00000062257
|
slc39a1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr19_+_9050852 | 0.11 |
ENSDART00000151031
|
ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila) |
| chr12_+_3723650 | 0.10 |
ENSDART00000179922
ENSDART00000152482 ENSDART00000108771 |
pagr1
|
PAXIP1 associated glutamate-rich protein 1 |
| chr7_+_31051213 | 0.10 |
ENSDART00000148347
|
tjp1a
|
tight junction protein 1a |
| chr20_-_2355357 | 0.10 |
ENSDART00000085281
|
EPB41L2
|
si:ch73-18b11.1 |
| chr23_-_270847 | 0.10 |
ENSDART00000191867
|
anks1aa
|
ankyrin repeat and sterile alpha motif domain containing 1Aa |
| chr4_-_890220 | 0.10 |
ENSDART00000022668
|
aim1b
|
crystallin beta-gamma domain containing 1b |
| chr1_+_58990121 | 0.10 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr23_+_43770149 | 0.10 |
ENSDART00000024313
|
rnf150b
|
ring finger protein 150b |
| chr23_+_2825940 | 0.10 |
ENSDART00000135781
|
plcg1
|
phospholipase C, gamma 1 |
| chr13_-_4367083 | 0.10 |
ENSDART00000124716
|
si:dkeyp-121d4.3
|
si:dkeyp-121d4.3 |
| chr19_+_47290287 | 0.10 |
ENSDART00000078382
|
tpmt.1
|
thiopurine S-methyltransferase, tandem duplicate 1 |
| chr21_+_233271 | 0.10 |
ENSDART00000171440
|
dtwd2
|
DTW domain containing 2 |
| chr2_-_39759059 | 0.10 |
ENSDART00000007333
|
slc25a36a
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36a |
| chr7_+_22809905 | 0.10 |
ENSDART00000166900
ENSDART00000143455 ENSDART00000126037 |
sf1
|
splicing factor 1 |
| chr10_-_1718395 | 0.09 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr22_+_17531020 | 0.09 |
ENSDART00000137689
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr22_-_17671348 | 0.09 |
ENSDART00000137995
|
tjp3
|
tight junction protein 3 |
| chr13_-_10431476 | 0.09 |
ENSDART00000133968
|
camkmt
|
calmodulin-lysine N-methyltransferase |
| chr1_-_22678471 | 0.09 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr5_+_63322093 | 0.09 |
ENSDART00000158086
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr10_-_35177257 | 0.09 |
ENSDART00000077426
|
pom121
|
POM121 transmembrane nucleoporin |
| chr3_-_16039619 | 0.09 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr7_+_39720317 | 0.09 |
ENSDART00000164996
|
si:dkey-58i8.5
|
si:dkey-58i8.5 |
| chr12_+_13348918 | 0.09 |
ENSDART00000181373
|
rnasen
|
ribonuclease type III, nuclear |
| chr16_-_28658341 | 0.09 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr21_+_3166988 | 0.09 |
ENSDART00000168163
|
hnrnpabb
|
heterogeneous nuclear ribonucleoprotein A/Bb |
| chr22_+_110158 | 0.09 |
ENSDART00000143698
|
prkar2ab
|
protein kinase, cAMP-dependent, regulatory, type II, alpha, B |
| chr11_+_12811906 | 0.09 |
ENSDART00000123445
|
rtel1
|
regulator of telomere elongation helicase 1 |
| chr6_-_1820606 | 0.09 |
ENSDART00000183228
|
FO834857.1
|
|
| chr19_+_770300 | 0.09 |
ENSDART00000062518
|
gstr
|
glutathione S-transferase rho |
| chr22_-_38274188 | 0.09 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr21_-_41369539 | 0.09 |
ENSDART00000187546
|
cpeb4b
|
cytoplasmic polyadenylation element binding protein 4b |
| chr25_+_258883 | 0.09 |
ENSDART00000155256
|
zgc:92481
|
zgc:92481 |
| chr23_+_2717950 | 0.08 |
ENSDART00000137641
|
ncoa6
|
nuclear receptor coactivator 6 |
| chr22_+_17531214 | 0.08 |
ENSDART00000161174
ENSDART00000088356 |
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| chr7_+_1550966 | 0.08 |
ENSDART00000177863
ENSDART00000126840 |
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr16_-_11798994 | 0.08 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr21_-_21790372 | 0.08 |
ENSDART00000151094
|
xrra1
|
X-ray radiation resistance associated 1 |
| chr19_+_4892281 | 0.08 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr13_-_26799244 | 0.08 |
ENSDART00000036419
|
vrk2
|
vaccinia related kinase 2 |
| chr20_-_147574 | 0.08 |
ENSDART00000104762
ENSDART00000131635 |
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr2_+_1989941 | 0.08 |
ENSDART00000190814
|
ssx2ipa
|
synovial sarcoma, X breakpoint 2 interacting protein a |
| chr19_-_82504 | 0.08 |
ENSDART00000027864
ENSDART00000160560 |
hnrnpr
|
heterogeneous nuclear ribonucleoprotein R |
| chr25_+_20116407 | 0.08 |
ENSDART00000183615
ENSDART00000193243 |
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr2_-_2642476 | 0.08 |
ENSDART00000124032
|
serbp1b
|
SERPINE1 mRNA binding protein 1b |
| chr11_+_45299447 | 0.08 |
ENSDART00000172999
|
mafgb
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Gb |
| chr3_-_32818607 | 0.08 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr7_-_20241346 | 0.07 |
ENSDART00000173619
ENSDART00000127699 |
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr22_+_396840 | 0.07 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr8_+_29749017 | 0.07 |
ENSDART00000185144
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr7_-_9674073 | 0.07 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr18_+_15106518 | 0.07 |
ENSDART00000168639
|
cry1ab
|
cryptochrome circadian clock 1ab |
| chr10_-_641609 | 0.07 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr3_-_36210344 | 0.07 |
ENSDART00000025326
|
csnk1da
|
casein kinase 1, delta a |
| chr3_-_38777553 | 0.07 |
ENSDART00000193045
|
znf281a
|
zinc finger protein 281a |
| chr3_-_34547000 | 0.07 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr24_+_32411753 | 0.07 |
ENSDART00000058530
|
neurod6a
|
neuronal differentiation 6a |
| chr11_+_31324335 | 0.07 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr23_-_14990865 | 0.07 |
ENSDART00000147799
|
ndrg3b
|
ndrg family member 3b |
| chr19_+_14352332 | 0.07 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr23_+_2740741 | 0.07 |
ENSDART00000134938
|
zgc:114123
|
zgc:114123 |
| chr3_+_14157090 | 0.07 |
ENSDART00000156766
|
si:ch211-108d22.2
|
si:ch211-108d22.2 |
| chr15_+_29123031 | 0.07 |
ENSDART00000133988
ENSDART00000060030 |
zgc:101731
|
zgc:101731 |
| chr22_-_7778895 | 0.07 |
ENSDART00000125489
|
si:ch73-44m9.3
|
si:ch73-44m9.3 |
| chr5_+_6670945 | 0.07 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr24_-_3407507 | 0.07 |
ENSDART00000132648
|
nck1b
|
NCK adaptor protein 1b |
| chr5_+_24087035 | 0.07 |
ENSDART00000183644
|
tp53
|
tumor protein p53 |
| chr3_-_34528306 | 0.07 |
ENSDART00000023039
|
sept9a
|
septin 9a |
| chr7_+_51795667 | 0.07 |
ENSDART00000174201
ENSDART00000073839 |
slc38a7
|
solute carrier family 38, member 7 |
| chr1_-_18446161 | 0.06 |
ENSDART00000089442
|
klhl5
|
kelch-like family member 5 |
| chr18_-_44356656 | 0.06 |
ENSDART00000193084
|
prdm10
|
PR domain containing 10 |
| chr16_-_32975951 | 0.06 |
ENSDART00000101969
ENSDART00000175149 |
me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr3_-_26204867 | 0.06 |
ENSDART00000103748
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr24_-_24060632 | 0.06 |
ENSDART00000090514
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr16_-_34258931 | 0.06 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr23_-_9965148 | 0.06 |
ENSDART00000137308
|
plxnb1a
|
plexin b1a |
| chr14_+_16287968 | 0.06 |
ENSDART00000106593
|
prpf19
|
pre-mRNA processing factor 19 |
| chr14_-_23801389 | 0.06 |
ENSDART00000054264
|
nr3c1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr25_+_2993855 | 0.06 |
ENSDART00000163009
|
neo1b
|
neogenin 1b |
| chr6_-_31233696 | 0.06 |
ENSDART00000079173
|
lepr
|
leptin receptor |
| chr15_+_35933094 | 0.06 |
ENSDART00000019976
|
rhbdd1
|
rhomboid domain containing 1 |
| chr2_+_13907452 | 0.06 |
ENSDART00000169724
ENSDART00000190691 |
zgc:66475
|
zgc:66475 |
| chr11_+_45343245 | 0.06 |
ENSDART00000160904
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr25_-_32449235 | 0.06 |
ENSDART00000115343
|
atp8b4
|
ATPase phospholipid transporting 8B4 |
| chr18_-_17087138 | 0.06 |
ENSDART00000135597
|
zc3h18
|
zinc finger CCCH-type containing 18 |
| chr11_-_44999858 | 0.06 |
ENSDART00000167759
ENSDART00000126845 |
ldb1b
|
LIM-domain binding 1b |
| chr11_+_13424116 | 0.06 |
ENSDART00000125563
|
homer3b
|
homer scaffolding protein 3b |
| chr24_-_24060460 | 0.06 |
ENSDART00000142813
|
abcc13
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| chr20_-_33675676 | 0.06 |
ENSDART00000147168
|
rock2b
|
rho-associated, coiled-coil containing protein kinase 2b |
| chr17_-_6613458 | 0.06 |
ENSDART00000175024
|
si:ch211-189e2.3
|
si:ch211-189e2.3 |
| chr18_+_13077800 | 0.06 |
ENSDART00000161153
|
GAN
|
gigaxonin |
| chr12_+_8003872 | 0.06 |
ENSDART00000180300
ENSDART00000126661 |
rhobtb1
|
Rho-related BTB domain containing 1 |
| chr17_-_38887424 | 0.06 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr8_+_45381298 | 0.06 |
ENSDART00000149451
ENSDART00000110364 |
tti2
|
TELO2 interacting protein 2 |
| chr11_-_44030962 | 0.06 |
ENSDART00000171910
|
FP016005.1
|
|
| chr24_-_25166720 | 0.05 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr10_-_42237304 | 0.05 |
ENSDART00000140341
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr19_+_48018802 | 0.05 |
ENSDART00000161339
ENSDART00000166978 |
UBE2M
|
si:ch1073-205c8.3 |
| chr9_+_2393764 | 0.05 |
ENSDART00000172624
|
chn1
|
chimerin 1 |
| chr2_+_23006792 | 0.05 |
ENSDART00000027782
|
mknk2a
|
MAP kinase interacting serine/threonine kinase 2a |
| chr4_+_77993977 | 0.05 |
ENSDART00000174118
|
terfa
|
telomeric repeat binding factor a |
| chr20_+_66857 | 0.05 |
ENSDART00000114999
|
lrfn2b
|
leucine rich repeat and fibronectin type III domain containing 2b |
| chr19_+_48060464 | 0.05 |
ENSDART00000123163
|
zgc:85936
|
zgc:85936 |
| chr15_-_33807758 | 0.05 |
ENSDART00000158445
|
pds5b
|
PDS5 cohesin associated factor B |
| chr24_+_24170914 | 0.05 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr17_+_43629008 | 0.05 |
ENSDART00000184185
ENSDART00000181681 |
znf365
|
zinc finger protein 365 |
| chr6_-_7726849 | 0.05 |
ENSDART00000151511
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr2_-_7185460 | 0.05 |
ENSDART00000092078
|
rc3h1b
|
ring finger and CCCH-type domains 1b |
| chr24_-_25166416 | 0.05 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr13_+_1100197 | 0.05 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| chr1_-_59232267 | 0.05 |
ENSDART00000169658
ENSDART00000163257 |
akap8l
|
A kinase (PRKA) anchor protein 8-like |
| chr12_-_2800809 | 0.05 |
ENSDART00000152682
ENSDART00000083784 |
ubtd1b
|
ubiquitin domain containing 1b |
| chr24_-_26715174 | 0.05 |
ENSDART00000079726
|
pld1b
|
phospholipase D1b |
| chr5_+_22098591 | 0.05 |
ENSDART00000143676
|
zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr16_-_23800484 | 0.05 |
ENSDART00000139964
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr19_-_45534392 | 0.05 |
ENSDART00000163920
|
trps1
|
trichorhinophalangeal syndrome I |
| chr13_+_33368503 | 0.05 |
ENSDART00000139650
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr25_-_21031007 | 0.05 |
ENSDART00000138985
|
gnaia
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide a |
| chr17_+_1514711 | 0.05 |
ENSDART00000165641
|
akt1
|
v-akt murine thymoma viral oncogene homolog 1 |
| chr8_+_54137350 | 0.04 |
ENSDART00000164153
|
brpf1
|
bromodomain and PHD finger containing, 1 |
| chr23_-_21957383 | 0.04 |
ENSDART00000145780
|
si:dkey-68l7.2
|
si:dkey-68l7.2 |
| chr19_-_2231146 | 0.04 |
ENSDART00000181909
ENSDART00000043595 |
twist1a
|
twist family bHLH transcription factor 1a |
| chr20_-_10487951 | 0.04 |
ENSDART00000064112
|
glrx5
|
glutaredoxin 5 homolog (S. cerevisiae) |
| chr16_-_42770064 | 0.04 |
ENSDART00000112879
|
slc4a7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr2_+_24867534 | 0.04 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr8_-_18613948 | 0.04 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr15_-_33527631 | 0.04 |
ENSDART00000187646
ENSDART00000161151 |
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr6_+_23809163 | 0.04 |
ENSDART00000170402
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr20_+_27646772 | 0.04 |
ENSDART00000141697
|
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr17_+_49081828 | 0.04 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr5_+_59234421 | 0.04 |
ENSDART00000045518
|
prkrip1
|
PRKR interacting protein 1 |
| chr15_-_434503 | 0.04 |
ENSDART00000122286
|
CABZ01056629.1
|
|
| chr5_+_3891485 | 0.04 |
ENSDART00000129329
ENSDART00000091711 |
rpain
|
RPA interacting protein |
| chr17_-_10059557 | 0.04 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr8_+_36554816 | 0.04 |
ENSDART00000126687
|
sf3a1
|
splicing factor 3a, subunit 1 |
| chr8_-_44611357 | 0.04 |
ENSDART00000063396
|
bag4
|
BCL2 associated athanogene 4 |
| chr10_-_26179805 | 0.04 |
ENSDART00000174797
|
trim3b
|
tripartite motif containing 3b |
| chr5_+_51102010 | 0.04 |
ENSDART00000110377
|
zgc:194398
|
zgc:194398 |
| chr12_+_35203091 | 0.04 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr24_+_27023616 | 0.04 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| chr14_-_36325558 | 0.04 |
ENSDART00000172783
|
egf
|
epidermal growth factor |
| chr5_-_32505276 | 0.04 |
ENSDART00000034705
ENSDART00000187597 |
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr24_-_26518972 | 0.04 |
ENSDART00000097792
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr7_+_73488035 | 0.04 |
ENSDART00000110330
|
arhgef40
|
Rho guanine nucleotide exchange factor (GEF) 40 |
| chr7_-_33829824 | 0.04 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr17_+_135590 | 0.04 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr11_-_25734417 | 0.04 |
ENSDART00000103570
|
brpf3a
|
bromodomain and PHD finger containing, 3a |
| chr19_+_49721 | 0.04 |
ENSDART00000160489
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr11_-_8782871 | 0.04 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr16_+_25285998 | 0.04 |
ENSDART00000154112
|
si:dkey-29h14.10
|
si:dkey-29h14.10 |
| chr5_-_31772559 | 0.04 |
ENSDART00000183879
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr7_-_25132592 | 0.04 |
ENSDART00000192214
|
badb
|
BCL2 associated agonist of cell death b |
| chr23_+_9057999 | 0.04 |
ENSDART00000091899
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr4_+_71401321 | 0.04 |
ENSDART00000158076
|
si:ch211-76m11.7
|
si:ch211-76m11.7 |
| chr1_+_31657842 | 0.03 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr1_+_16574312 | 0.03 |
ENSDART00000187067
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr1_-_31657854 | 0.03 |
ENSDART00000085309
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr4_-_2868112 | 0.03 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr6_+_23809501 | 0.03 |
ENSDART00000168701
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr7_-_29021757 | 0.03 |
ENSDART00000086905
|
nrn1lb
|
neuritin 1-like b |
| chr7_+_528593 | 0.03 |
ENSDART00000091955
|
nrxn2b
|
neurexin 2b |
| chr14_-_237130 | 0.03 |
ENSDART00000164988
|
bod1l1
|
biorientation of chromosomes in cell division 1-like 1 |
| chr20_+_54333774 | 0.03 |
ENSDART00000144633
|
cipcb
|
CLOCK-interacting pacemaker b |
| chr20_+_27647436 | 0.03 |
ENSDART00000180557
ENSDART00000185628 |
mthfd1a
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1a, methenyltetrahydrofolate cyclohydrolase, formyltetrahydrofolate synthetase |
| chr10_-_35236949 | 0.03 |
ENSDART00000145804
|
ypel2a
|
yippee-like 2a |
| chr11_+_37216668 | 0.03 |
ENSDART00000173076
|
zgc:112265
|
zgc:112265 |
| chr7_+_13491452 | 0.03 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr15_+_47418565 | 0.03 |
ENSDART00000155709
|
clpb
|
ClpB homolog, mitochondrial AAA ATPase chaperonin |
Network of associatons between targets according to the STRING database.
First level regulatory network of rcor1+rcor2+rcor3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035477 | regulation of angioblast cell migration involved in selective angioblast sprouting(GO:0035477) |
| 0.0 | 0.3 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0010610 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of cell junction assembly(GO:1901890) |
| 0.0 | 0.2 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.1 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.1 | GO:1904983 | transmembrane glycine transport from cytosol to mitochondrion(GO:1904983) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0032205 | negative regulation of telomere maintenance(GO:0032205) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.0 | 0.0 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0071548 | cellular response to cortisol stimulus(GO:0071387) response to dexamethasone(GO:0071548) |
| 0.0 | 0.1 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.1 | GO:1903723 | negative regulation of centriole elongation(GO:1903723) |
| 0.0 | 0.0 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.1 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.0 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.0 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.1 | GO:0010456 | cell proliferation in dorsal spinal cord(GO:0010456) |
| 0.0 | 0.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.1 | GO:0030104 | water homeostasis(GO:0030104) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0008119 | thiopurine S-methyltransferase activity(GO:0008119) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0015562 | protein-transmembrane transporting ATPase activity(GO:0015462) efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.0 | 0.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |