Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
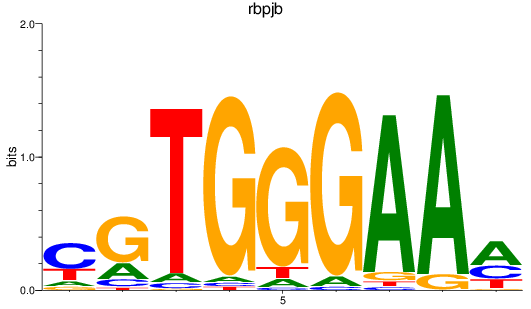
Results for rbpjb
Z-value: 0.67
Transcription factors associated with rbpjb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpjb
|
ENSDARG00000052091 | recombination signal binding protein for immunoglobulin kappa J region b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpjb | dr11_v1_chr7_+_62080456_62080456 | 0.80 | 5.7e-05 | Click! |
Activity profile of rbpjb motif
Sorted Z-values of rbpjb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_18899427 | 1.63 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr1_+_14454663 | 1.42 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr1_-_31105376 | 1.29 |
ENSDART00000132466
|
ppp1r9alb
|
protein phosphatase 1 regulatory subunit 9A-like B |
| chr6_+_10333920 | 1.14 |
ENSDART00000151667
ENSDART00000151477 |
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr21_-_13690712 | 1.10 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr13_-_33654931 | 1.09 |
ENSDART00000020350
|
snx5
|
sorting nexin 5 |
| chr20_+_14789148 | 1.03 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr19_-_27830818 | 0.93 |
ENSDART00000131767
|
papd7
|
PAP associated domain containing 7 |
| chr15_-_31147301 | 0.91 |
ENSDART00000157145
ENSDART00000155473 ENSDART00000048103 |
ksr1b
|
kinase suppressor of ras 1b |
| chr5_-_23675222 | 0.86 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr14_+_28486213 | 0.83 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr6_+_3716666 | 0.83 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr2_+_2110271 | 0.83 |
ENSDART00000148460
|
dspa
|
desmoplakin a |
| chr6_-_53144336 | 0.80 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr4_-_13613148 | 0.77 |
ENSDART00000067164
ENSDART00000111247 |
irf5
|
interferon regulatory factor 5 |
| chr21_-_4849029 | 0.77 |
ENSDART00000168930
ENSDART00000151019 |
notch1a
|
notch 1a |
| chr20_+_14789305 | 0.75 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr8_+_14915332 | 0.74 |
ENSDART00000164385
|
abl2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr19_-_25113660 | 0.73 |
ENSDART00000035538
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_-_18613948 | 0.73 |
ENSDART00000089172
|
cpox
|
coproporphyrinogen oxidase |
| chr7_+_62080456 | 0.70 |
ENSDART00000092580
|
rbpjb
|
recombination signal binding protein for immunoglobulin kappa J region b |
| chr16_+_33953644 | 0.70 |
ENSDART00000164447
ENSDART00000159969 |
arid1aa
|
AT rich interactive domain 1Aa (SWI-like) |
| chr9_-_23807032 | 0.68 |
ENSDART00000027443
|
esyt3
|
extended synaptotagmin-like protein 3 |
| chr21_-_27213166 | 0.68 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr11_+_34921492 | 0.62 |
ENSDART00000128070
|
gnai2a
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2a |
| chr6_-_9695294 | 0.61 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr20_-_32045057 | 0.60 |
ENSDART00000152970
ENSDART00000034248 |
rab32a
|
RAB32a, member RAS oncogene family |
| chr18_+_14277003 | 0.60 |
ENSDART00000006628
|
zgc:173742
|
zgc:173742 |
| chr18_+_22220656 | 0.59 |
ENSDART00000191862
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr23_-_36303216 | 0.59 |
ENSDART00000188720
|
cbx5
|
chromobox homolog 5 (HP1 alpha homolog, Drosophila) |
| chr21_+_31253048 | 0.57 |
ENSDART00000178521
ENSDART00000132317 ENSDART00000040190 |
asl
|
argininosuccinate lyase |
| chr9_+_2020667 | 0.57 |
ENSDART00000157818
|
lnpa
|
limb and neural patterns a |
| chr3_-_34547000 | 0.56 |
ENSDART00000166623
|
sept9a
|
septin 9a |
| chr20_-_38801981 | 0.55 |
ENSDART00000125333
|
cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr12_+_38563073 | 0.55 |
ENSDART00000009172
|
ttyh2
|
tweety family member 2 |
| chr24_-_6375774 | 0.54 |
ENSDART00000013294
|
anxa13
|
annexin A13 |
| chr4_-_4250317 | 0.54 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr19_-_11015238 | 0.53 |
ENSDART00000010997
|
tpm3
|
tropomyosin 3 |
| chr5_+_25736979 | 0.53 |
ENSDART00000175959
|
abhd17b
|
abhydrolase domain containing 17B |
| chr8_+_21225064 | 0.52 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr23_-_21758253 | 0.52 |
ENSDART00000046613
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr1_+_10018466 | 0.51 |
ENSDART00000113551
|
trim2b
|
tripartite motif containing 2b |
| chr18_-_16953978 | 0.50 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr22_-_11054244 | 0.50 |
ENSDART00000105823
|
insrb
|
insulin receptor b |
| chr7_+_52712807 | 0.48 |
ENSDART00000174095
ENSDART00000174377 ENSDART00000174061 ENSDART00000174094 ENSDART00000110906 ENSDART00000174071 ENSDART00000174238 |
znf280d
|
zinc finger protein 280D |
| chr1_-_46924801 | 0.48 |
ENSDART00000142560
|
pdxkb
|
pyridoxal (pyridoxine, vitamin B6) kinase b |
| chr13_-_31389661 | 0.48 |
ENSDART00000134630
|
zdhhc16a
|
zinc finger, DHHC-type containing 16a |
| chr9_+_19623363 | 0.46 |
ENSDART00000142471
ENSDART00000147662 ENSDART00000136053 |
pdxka
|
pyridoxal (pyridoxine, vitamin B6) kinase a |
| chr5_-_36597612 | 0.46 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr13_-_36798204 | 0.46 |
ENSDART00000012357
|
sav1
|
salvador family WW domain containing protein 1 |
| chr19_+_40248697 | 0.45 |
ENSDART00000151269
|
cdk6
|
cyclin-dependent kinase 6 |
| chr2_-_57941037 | 0.44 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr3_+_31058464 | 0.44 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr15_-_47468085 | 0.43 |
ENSDART00000164438
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr20_+_45620076 | 0.43 |
ENSDART00000113806
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr5_+_33287611 | 0.42 |
ENSDART00000125093
ENSDART00000146759 |
med22
|
mediator complex subunit 22 |
| chr10_+_17026870 | 0.42 |
ENSDART00000184529
ENSDART00000157480 |
CR855996.2
|
|
| chr3_+_46315016 | 0.42 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr14_+_49152341 | 0.42 |
ENSDART00000084114
|
nsd1a
|
nuclear receptor binding SET domain protein 1a |
| chr8_+_48947608 | 0.41 |
ENSDART00000184742
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr25_+_20272145 | 0.39 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr17_+_49081828 | 0.39 |
ENSDART00000156492
|
tiam2a
|
T cell lymphoma invasion and metastasis 2a |
| chr21_+_43485512 | 0.38 |
ENSDART00000026666
|
atg4a
|
autophagy related 4A, cysteine peptidase |
| chr16_-_21047872 | 0.38 |
ENSDART00000131582
|
cbx3b
|
chromobox homolog 3b |
| chr11_-_11518469 | 0.37 |
ENSDART00000104254
|
krt15
|
keratin 15 |
| chr19_-_7540821 | 0.37 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr23_-_36003441 | 0.37 |
ENSDART00000164699
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr15_+_20530649 | 0.36 |
ENSDART00000186312
|
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr3_+_27722355 | 0.36 |
ENSDART00000132761
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr24_+_31374324 | 0.35 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr25_-_893464 | 0.35 |
ENSDART00000159321
|
znf609a
|
zinc finger protein 609a |
| chr23_-_36003282 | 0.35 |
ENSDART00000103150
|
calcoco1a
|
calcium binding and coiled-coil domain 1a |
| chr5_+_61467618 | 0.35 |
ENSDART00000074073
ENSDART00000182094 |
alkbh4
|
alkB homolog 4, lysine demthylase |
| chr7_+_27834130 | 0.35 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr6_+_30430591 | 0.34 |
ENSDART00000108943
|
shroom2a
|
shroom family member 2a |
| chr24_+_28953089 | 0.34 |
ENSDART00000153761
|
rnpc3
|
RNA-binding region (RNP1, RRM) containing 3 |
| chr13_+_15657911 | 0.33 |
ENSDART00000134972
ENSDART00000138991 ENSDART00000133342 |
mark3a
|
MAP/microtubule affinity-regulating kinase 3a |
| chr13_-_24874950 | 0.33 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr12_-_13729263 | 0.32 |
ENSDART00000078021
|
foxh1
|
forkhead box H1 |
| chr12_+_1286642 | 0.32 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr16_-_21047483 | 0.32 |
ENSDART00000136235
|
cbx3b
|
chromobox homolog 3b |
| chr23_+_32011768 | 0.31 |
ENSDART00000053509
|
plagx
|
pleiomorphic adenoma gene X |
| chr20_+_22799857 | 0.31 |
ENSDART00000058527
|
scfd2
|
sec1 family domain containing 2 |
| chr13_-_33700461 | 0.30 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr13_-_33246154 | 0.30 |
ENSDART00000130127
|
rtf1
|
RTF1 homolog, Paf1/RNA polymerase II complex component |
| chr10_+_35329751 | 0.30 |
ENSDART00000148043
|
si:dkey-259j3.5
|
si:dkey-259j3.5 |
| chr3_-_41995321 | 0.29 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr5_-_22082918 | 0.29 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr20_+_22799641 | 0.28 |
ENSDART00000131132
|
scfd2
|
sec1 family domain containing 2 |
| chr10_-_42297889 | 0.28 |
ENSDART00000099262
|
tcf7l1a
|
transcription factor 7 like 1a |
| chr8_+_26396552 | 0.28 |
ENSDART00000087151
|
amt
|
aminomethyltransferase |
| chr24_-_20658446 | 0.28 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr5_+_20035284 | 0.27 |
ENSDART00000191808
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr6_-_12172424 | 0.27 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr14_+_41406321 | 0.27 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr23_+_25172682 | 0.27 |
ENSDART00000191197
ENSDART00000183497 |
si:dkey-151g10.3
|
si:dkey-151g10.3 |
| chr8_-_11067079 | 0.27 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr25_+_6122823 | 0.25 |
ENSDART00000191824
ENSDART00000067514 |
rbpms2a
|
RNA binding protein with multiple splicing 2a |
| chr19_+_24896409 | 0.24 |
ENSDART00000049840
|
eya3
|
EYA transcriptional coactivator and phosphatase 3 |
| chr18_+_15644559 | 0.24 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr25_+_35304903 | 0.24 |
ENSDART00000034313
ENSDART00000187659 |
gas2a
|
growth arrest-specific 2a |
| chr8_-_48847772 | 0.24 |
ENSDART00000122458
|
wrap73
|
WD repeat containing, antisense to TP73 |
| chr24_-_19718077 | 0.24 |
ENSDART00000109107
ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr21_-_40562705 | 0.24 |
ENSDART00000158289
ENSDART00000171997 |
taok1b
|
TAO kinase 1b |
| chr9_+_38481780 | 0.23 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr16_-_9869056 | 0.23 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr2_+_31957554 | 0.23 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr21_-_43485351 | 0.23 |
ENSDART00000151597
|
ankrd46a
|
ankyrin repeat domain 46a |
| chr19_+_40322912 | 0.23 |
ENSDART00000146893
|
cdk6
|
cyclin-dependent kinase 6 |
| chr17_-_49339677 | 0.23 |
ENSDART00000047857
|
orc3
|
origin recognition complex, subunit 3 |
| chr2_+_1881334 | 0.22 |
ENSDART00000161420
|
adgrl2b.1
|
adhesion G protein-coupled receptor L2b, tandem duplicate 1 |
| chr13_-_31017960 | 0.22 |
ENSDART00000145287
|
wdfy4
|
WDFY family member 4 |
| chr22_+_12431608 | 0.22 |
ENSDART00000108609
|
rnd3a
|
Rho family GTPase 3a |
| chr25_-_14637660 | 0.21 |
ENSDART00000143666
|
nav2b
|
neuron navigator 2b |
| chr2_-_898899 | 0.21 |
ENSDART00000058289
|
dusp22b
|
dual specificity phosphatase 22b |
| chr1_+_1941031 | 0.20 |
ENSDART00000110331
|
PTGFRN
|
si:ch211-132g1.7 |
| chr3_+_35608385 | 0.20 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr8_-_15150131 | 0.19 |
ENSDART00000138253
|
bcar3
|
BCAR3, NSP family adaptor protein |
| chr6_-_14292307 | 0.18 |
ENSDART00000177852
ENSDART00000061745 |
inpp4ab
|
inositol polyphosphate-4-phosphatase type I Ab |
| chr16_-_13881139 | 0.18 |
ENSDART00000138904
ENSDART00000090221 ENSDART00000135099 |
cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr24_-_24796583 | 0.18 |
ENSDART00000144791
ENSDART00000146570 |
pde7a
|
phosphodiesterase 7A |
| chr5_-_2112030 | 0.17 |
ENSDART00000091932
|
gusb
|
glucuronidase, beta |
| chr6_+_55006686 | 0.17 |
ENSDART00000081703
|
nav1b
|
neuron navigator 1b |
| chr13_+_11440389 | 0.16 |
ENSDART00000186463
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr4_+_4849789 | 0.16 |
ENSDART00000130818
ENSDART00000127751 |
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr4_-_15103646 | 0.15 |
ENSDART00000138183
ENSDART00000181044 |
nrf1
|
nuclear respiratory factor 1 |
| chr5_-_69312533 | 0.15 |
ENSDART00000082614
ENSDART00000183098 |
smtnb
|
smoothelin b |
| chr22_+_20141528 | 0.15 |
ENSDART00000187770
|
eef2a.1
|
eukaryotic translation elongation factor 2a, tandem duplicate 1 |
| chr6_+_58522557 | 0.15 |
ENSDART00000128062
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr18_+_6558146 | 0.15 |
ENSDART00000169401
|
b4galnt3b
|
beta-1,4-N-acetyl-galactosaminyl transferase 3b |
| chr19_-_12648408 | 0.15 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr25_+_10793478 | 0.15 |
ENSDART00000058339
ENSDART00000134923 |
ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr7_-_15185811 | 0.14 |
ENSDART00000031049
|
si:dkey-172h23.2
|
si:dkey-172h23.2 |
| chr6_+_58522738 | 0.14 |
ENSDART00000157327
|
arfrp1
|
ADP-ribosylation factor related protein 1 |
| chr6_+_21062416 | 0.14 |
ENSDART00000141643
|
si:dkey-91f15.1
|
si:dkey-91f15.1 |
| chr5_+_62009765 | 0.14 |
ENSDART00000134335
|
akap10
|
A kinase (PRKA) anchor protein 10 |
| chr12_+_21299338 | 0.13 |
ENSDART00000074540
ENSDART00000133188 |
ca10a
|
carbonic anhydrase Xa |
| chr6_-_25384526 | 0.13 |
ENSDART00000160544
|
PKN2 (1 of many)
|
zgc:153916 |
| chr14_+_23076207 | 0.12 |
ENSDART00000161628
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr25_+_6186823 | 0.11 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr17_-_27235797 | 0.11 |
ENSDART00000130080
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr7_+_38770167 | 0.11 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr5_-_25072607 | 0.11 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr4_+_74943111 | 0.11 |
ENSDART00000004739
|
nup50
|
nucleoporin 50 |
| chr5_+_62009930 | 0.10 |
ENSDART00000082872
|
akap10
|
A kinase (PRKA) anchor protein 10 |
| chr1_-_51720633 | 0.10 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr3_-_40136743 | 0.09 |
ENSDART00000149546
|
myo15aa
|
myosin XVAa |
| chr18_+_26829086 | 0.09 |
ENSDART00000098356
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr6_-_25384792 | 0.09 |
ENSDART00000169030
|
PKN2 (1 of many)
|
zgc:153916 |
| chr17_-_27223965 | 0.09 |
ENSDART00000192577
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr17_+_24851951 | 0.09 |
ENSDART00000180746
|
cx35.4
|
connexin 35.4 |
| chr11_-_1409236 | 0.08 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr11_-_27962757 | 0.08 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr5_+_38837429 | 0.08 |
ENSDART00000160236
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr7_-_26436436 | 0.08 |
ENSDART00000019035
ENSDART00000123395 |
her8a
|
hairy-related 8a |
| chr11_-_7320211 | 0.08 |
ENSDART00000091664
|
apc2
|
adenomatosis polyposis coli 2 |
| chr8_-_13010011 | 0.08 |
ENSDART00000140969
|
dennd2da
|
DENN/MADD domain containing 2Da |
| chr6_+_39923052 | 0.08 |
ENSDART00000149019
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr9_-_12574473 | 0.08 |
ENSDART00000191372
ENSDART00000193667 |
igf2bp2a
|
insulin-like growth factor 2 mRNA binding protein 2a |
| chr18_-_11184584 | 0.06 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr9_-_7089303 | 0.06 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr17_-_27200634 | 0.05 |
ENSDART00000185332
|
asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr7_+_53754653 | 0.05 |
ENSDART00000163261
ENSDART00000158160 |
neo1a
|
neogenin 1a |
| chr3_+_59051503 | 0.05 |
ENSDART00000160767
|
rasd4
|
rasd family member 4 |
| chr19_-_12648122 | 0.05 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr7_+_53755054 | 0.05 |
ENSDART00000181629
|
neo1a
|
neogenin 1a |
| chr18_+_26829362 | 0.05 |
ENSDART00000132728
|
slc28a1
|
solute carrier family 28 (concentrative nucleoside transporter), member 1 |
| chr1_+_54683655 | 0.04 |
ENSDART00000132785
|
knop1
|
lysine-rich nucleolar protein 1 |
| chr10_+_3507861 | 0.04 |
ENSDART00000092684
|
rph3aa
|
rabphilin 3A homolog (mouse), a |
| chr16_-_52646789 | 0.04 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr8_+_21159122 | 0.04 |
ENSDART00000033491
|
spryd4
|
SPRY domain containing 4 |
| chr1_-_40016058 | 0.04 |
ENSDART00000165373
|
cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr9_-_22205682 | 0.03 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr21_-_2621127 | 0.03 |
ENSDART00000172363
|
col4a3bpb
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein b |
| chr19_-_32641725 | 0.03 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr21_+_20549395 | 0.03 |
ENSDART00000181633
|
efna5a
|
ephrin-A5a |
| chr9_+_7358749 | 0.02 |
ENSDART00000081660
|
ihha
|
Indian hedgehog homolog a |
| chr14_-_32503363 | 0.02 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr25_-_29072162 | 0.02 |
ENSDART00000169269
|
arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr11_-_29563437 | 0.02 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr7_-_26462831 | 0.01 |
ENSDART00000113543
|
mblac1
|
metallo-beta-lactamase domain containing 1 |
| chr9_-_22076368 | 0.01 |
ENSDART00000128486
|
crygm2a
|
crystallin, gamma M2a |
| chr15_+_26603395 | 0.00 |
ENSDART00000188667
|
slc47a3
|
solute carrier family 47 (multidrug and toxin extrusion), member 3 |
| chr5_+_44944778 | 0.00 |
ENSDART00000130428
ENSDART00000044361 ENSDART00000128825 ENSDART00000124637 ENSDART00000126066 ENSDART00000177635 |
dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr4_+_62598975 | 0.00 |
ENSDART00000163548
|
si:ch211-79g12.2
|
si:ch211-79g12.2 |
| chr13_+_33655404 | 0.00 |
ENSDART00000023379
|
mgme1
|
mitochondrial genome maintenance exonuclease 1 |
| chr7_-_33960170 | 0.00 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpjb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.3 | 0.9 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.3 | 0.8 | GO:0021531 | spinal cord radial glial cell differentiation(GO:0021531) |
| 0.2 | 1.8 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.2 | 0.6 | GO:1903373 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of endoplasmic reticulum tubular network organization(GO:1903373) |
| 0.2 | 0.5 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.1 | 0.6 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.1 | 1.4 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.1 | 0.3 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.1 | 0.5 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.5 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.3 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.3 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 0.6 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.3 | GO:0051145 | smooth muscle cell differentiation(GO:0051145) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.3 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.5 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.5 | GO:0051897 | positive regulation of protein kinase B signaling(GO:0051897) |
| 0.0 | 0.3 | GO:0008105 | asymmetric protein localization(GO:0008105) apical protein localization(GO:0045176) |
| 0.0 | 0.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0071623 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.3 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.2 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.5 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.8 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 1.1 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.2 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.0 | 0.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.3 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.3 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.8 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.2 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0072698 | protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.8 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.4 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0098826 | endoplasmic reticulum tubular network membrane(GO:0098826) |
| 0.1 | 0.6 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 0.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.7 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 1.8 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.8 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.4 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0005922 | connexon complex(GO:0005922) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.6 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.5 | GO:0043560 | insulin-activated receptor activity(GO:0005009) insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.7 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.1 | 0.7 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.7 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.0 | 1.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 1.3 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.2 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.3 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.0 | 0.8 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.5 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 2.4 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 1.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 1.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |