Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
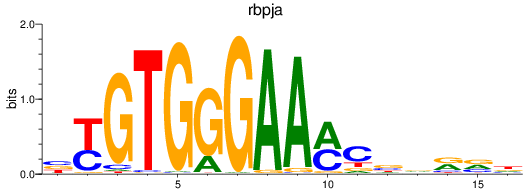
Results for rbpja
Z-value: 0.53
Transcription factors associated with rbpja
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rbpja
|
ENSDARG00000003398 | recombination signal binding protein for immunoglobulin kappa J region a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rbpja | dr11_v1_chr1_+_14454663_14454663 | -0.30 | 2.3e-01 | Click! |
Activity profile of rbpja motif
Sorted Z-values of rbpja motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_2573021 | 1.97 |
ENSDART00000074181
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr24_-_9989634 | 1.89 |
ENSDART00000115275
|
zgc:152652
|
zgc:152652 |
| chr1_-_58887610 | 1.67 |
ENSDART00000180647
|
CABZ01084501.1
|
microfibril-associated glycoprotein 4-like precursor |
| chr3_-_3428938 | 1.42 |
ENSDART00000179811
ENSDART00000115282 ENSDART00000192263 ENSDART00000046454 |
A2ML1 (1 of many)
|
zgc:171445 |
| chr3_-_3439150 | 1.22 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr6_+_21005725 | 0.86 |
ENSDART00000041370
|
cx44.2
|
connexin 44.2 |
| chr2_-_38287987 | 0.79 |
ENSDART00000185329
ENSDART00000061677 |
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr20_+_14789148 | 0.76 |
ENSDART00000164761
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr17_+_16046132 | 0.71 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr5_-_32338866 | 0.62 |
ENSDART00000017956
ENSDART00000047670 |
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr19_-_18127808 | 0.60 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr17_+_11372531 | 0.60 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr19_+_7001170 | 0.58 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr19_+_42432625 | 0.57 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr23_-_27571667 | 0.56 |
ENSDART00000008174
|
pfkma
|
phosphofructokinase, muscle a |
| chr11_-_3535537 | 0.55 |
ENSDART00000165329
ENSDART00000009788 |
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr25_+_2263857 | 0.53 |
ENSDART00000076439
|
yars2
|
tyrosyl-tRNA synthetase 2, mitochondrial |
| chr19_-_18127629 | 0.52 |
ENSDART00000187722
|
snx10a
|
sorting nexin 10a |
| chr2_+_8112449 | 0.51 |
ENSDART00000138136
|
chst2a
|
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2a |
| chr17_+_45472608 | 0.51 |
ENSDART00000103493
ENSDART00000183474 |
zgc:110783
|
zgc:110783 |
| chr3_-_53092509 | 0.51 |
ENSDART00000062081
|
lpar2a
|
lysophosphatidic acid receptor 2a |
| chr6_-_53144336 | 0.50 |
ENSDART00000154429
|
gnai2b
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2b |
| chr21_+_39673157 | 0.50 |
ENSDART00000100240
|
mrm3b
|
mitochondrial rRNA methyltransferase 3b |
| chr18_+_18612388 | 0.48 |
ENSDART00000186455
|
st3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr5_+_36850650 | 0.48 |
ENSDART00000051186
|
nccrp1
|
non-specific cytotoxic cell receptor protein 1 |
| chr16_-_25680666 | 0.43 |
ENSDART00000132693
ENSDART00000140539 ENSDART00000015302 |
tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr21_-_32082130 | 0.42 |
ENSDART00000003978
ENSDART00000182050 |
mat2b
|
methionine adenosyltransferase II, beta |
| chr5_-_36597612 | 0.41 |
ENSDART00000031270
ENSDART00000122098 |
rhogc
|
ras homolog gene family, member Gc |
| chr23_-_5783421 | 0.41 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr18_-_21746421 | 0.40 |
ENSDART00000188809
|
pskh1
|
protein serine kinase H1 |
| chr20_+_14789305 | 0.39 |
ENSDART00000002463
|
tmed5
|
transmembrane p24 trafficking protein 5 |
| chr15_-_28908027 | 0.39 |
ENSDART00000182790
ENSDART00000192461 |
eml2
|
echinoderm microtubule associated protein like 2 |
| chr10_-_38468847 | 0.39 |
ENSDART00000133914
|
gdpd5a
|
glycerophosphodiester phosphodiesterase domain containing 5a |
| chr7_-_32875552 | 0.38 |
ENSDART00000132504
|
ano5b
|
anoctamin 5b |
| chr15_-_28907709 | 0.38 |
ENSDART00000017268
|
eml2
|
echinoderm microtubule associated protein like 2 |
| chr17_+_5061135 | 0.38 |
ENSDART00000064313
|
cdc5l
|
CDC5 cell division cycle 5-like (S. pombe) |
| chr24_+_7884880 | 0.38 |
ENSDART00000139467
|
bmp6
|
bone morphogenetic protein 6 |
| chr19_-_12648122 | 0.37 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr21_+_12010505 | 0.37 |
ENSDART00000123522
|
aqp7
|
aquaporin 7 |
| chr3_+_36646054 | 0.36 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr17_+_43659940 | 0.36 |
ENSDART00000145738
ENSDART00000075619 |
adob
|
2-aminoethanethiol (cysteamine) dioxygenase b |
| chr3_+_35608385 | 0.36 |
ENSDART00000193219
ENSDART00000132703 |
traf7
|
TNF receptor-associated factor 7 |
| chr21_+_22833905 | 0.35 |
ENSDART00000111150
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr5_+_65040228 | 0.35 |
ENSDART00000164278
|
pmpca
|
peptidase (mitochondrial processing) alpha |
| chr19_+_3842891 | 0.35 |
ENSDART00000159043
|
lsm10
|
LSM10, U7 small nuclear RNA associated |
| chr14_-_33521071 | 0.35 |
ENSDART00000052789
|
c1galt1c1
|
C1GALT1-specific chaperone 1 |
| chr13_+_21919786 | 0.33 |
ENSDART00000182440
|
ndst2a
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2a |
| chr15_-_25435085 | 0.33 |
ENSDART00000112079
|
tlcd2
|
TLC domain containing 2 |
| chr18_+_43183749 | 0.33 |
ENSDART00000151166
|
nectin1b
|
nectin cell adhesion molecule 1b |
| chr8_-_46386024 | 0.31 |
ENSDART00000136602
ENSDART00000060919 ENSDART00000137472 |
qars
|
glutaminyl-tRNA synthetase |
| chr16_+_8139515 | 0.31 |
ENSDART00000193303
ENSDART00000139432 |
pomgnt2
|
protein O-linked mannose N-acetylglucosaminyltransferase 2 (beta 1,4-) |
| chr20_+_53368611 | 0.30 |
ENSDART00000060432
|
cdc40
|
cell division cycle 40 homolog (S. cerevisiae) |
| chr18_-_21640389 | 0.29 |
ENSDART00000100857
|
slc38a8a
|
solute carrier family 38, member 8a |
| chr17_-_6508406 | 0.29 |
ENSDART00000002778
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
| chr25_-_893464 | 0.28 |
ENSDART00000159321
|
znf609a
|
zinc finger protein 609a |
| chr13_-_49819027 | 0.28 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr3_+_32862730 | 0.28 |
ENSDART00000144939
ENSDART00000125126 ENSDART00000144531 ENSDART00000141717 ENSDART00000137599 |
zgc:162613
|
zgc:162613 |
| chr21_-_2209012 | 0.28 |
ENSDART00000158345
|
zgc:162971
|
zgc:162971 |
| chr16_+_2905150 | 0.28 |
ENSDART00000109980
|
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr6_+_52235441 | 0.27 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr8_+_47804284 | 0.26 |
ENSDART00000053285
|
ndufa7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 |
| chr6_-_9695294 | 0.26 |
ENSDART00000162728
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr1_-_51720633 | 0.26 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr7_+_24496894 | 0.26 |
ENSDART00000149994
|
nelfa
|
negative elongation factor complex member A |
| chr11_-_26701611 | 0.26 |
ENSDART00000083010
|
acad9
|
acyl-CoA dehydrogenase family, member 9 |
| chr11_-_27537593 | 0.25 |
ENSDART00000173444
ENSDART00000172895 ENSDART00000088177 |
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr15_-_31147301 | 0.24 |
ENSDART00000157145
ENSDART00000155473 ENSDART00000048103 |
ksr1b
|
kinase suppressor of ras 1b |
| chr23_-_478201 | 0.22 |
ENSDART00000140749
|
si:ch73-181d5.4
|
si:ch73-181d5.4 |
| chr20_+_5065268 | 0.22 |
ENSDART00000053883
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr11_+_23957440 | 0.22 |
ENSDART00000190721
|
cntn2
|
contactin 2 |
| chr5_+_27137473 | 0.22 |
ENSDART00000181833
|
unc5db
|
unc-5 netrin receptor Db |
| chr20_-_54075136 | 0.21 |
ENSDART00000074255
|
mgat2
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr14_+_32926385 | 0.21 |
ENSDART00000139159
|
lnx2b
|
ligand of numb-protein X 2b |
| chr19_+_13994563 | 0.20 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr24_+_30120815 | 0.20 |
ENSDART00000157979
|
plppr5b
|
phospholipid phosphatase related 5b |
| chr6_-_55423220 | 0.20 |
ENSDART00000158929
|
ctsa
|
cathepsin A |
| chr2_-_41518340 | 0.20 |
ENSDART00000130830
|
otomp
|
otolith matrix protein |
| chr6_+_29860776 | 0.20 |
ENSDART00000028406
|
dlg1
|
discs, large homolog 1 (Drosophila) |
| chr5_+_20319519 | 0.19 |
ENSDART00000004217
|
coro1ca
|
coronin, actin binding protein, 1Ca |
| chr21_+_26536950 | 0.19 |
ENSDART00000146315
|
stx5al
|
syntaxin 5A, like |
| chr18_-_18587745 | 0.19 |
ENSDART00000191973
|
sf3b3
|
splicing factor 3b, subunit 3 |
| chr5_+_4533244 | 0.19 |
ENSDART00000158826
|
CABZ01058650.1
|
Danio rerio thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 (LOC561325), mRNA. |
| chr19_-_12648408 | 0.19 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr8_+_15277874 | 0.19 |
ENSDART00000146965
|
dnttip2
|
deoxynucleotidyltransferase, terminal, interacting protein 2 |
| chr10_-_36808348 | 0.19 |
ENSDART00000099320
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr5_-_23675222 | 0.19 |
ENSDART00000135153
|
TBC1D8B
|
si:dkey-110k5.6 |
| chr9_-_7089303 | 0.18 |
ENSDART00000146609
|
coa5
|
cytochrome C oxidase assembly factor 5 |
| chr16_-_27161410 | 0.18 |
ENSDART00000177503
|
LO017771.1
|
|
| chr17_+_22381215 | 0.18 |
ENSDART00000162670
ENSDART00000128875 |
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr14_-_4321874 | 0.18 |
ENSDART00000042672
|
guf1
|
GUF1 homolog, GTPase |
| chr17_+_44756247 | 0.18 |
ENSDART00000153773
|
cipca
|
CLOCK-interacting pacemaker a |
| chr17_+_5611166 | 0.17 |
ENSDART00000097455
|
fam167aa
|
family with sequence similarity 167, member Aa |
| chr2_-_32237916 | 0.17 |
ENSDART00000141418
|
fam49ba
|
family with sequence similarity 49, member Ba |
| chr10_+_573667 | 0.17 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr19_-_42462491 | 0.17 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr9_-_56399699 | 0.17 |
ENSDART00000170281
|
rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr2_-_43850897 | 0.16 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr18_-_50676567 | 0.16 |
ENSDART00000172264
|
ube2q1
|
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr12_-_37449396 | 0.16 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr18_-_46369516 | 0.16 |
ENSDART00000018163
|
irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr6_+_3716666 | 0.16 |
ENSDART00000041627
|
ssb
|
Sjogren syndrome antigen B (autoantigen La) |
| chr13_+_24552254 | 0.15 |
ENSDART00000147907
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr7_-_41726657 | 0.15 |
ENSDART00000099121
|
arl8
|
ADP-ribosylation factor-like 8 |
| chr11_+_15480695 | 0.15 |
ENSDART00000104111
|
cdk5rap1
|
CDK5 regulatory subunit associated protein 1 |
| chr21_+_43172506 | 0.14 |
ENSDART00000121725
|
zcchc10
|
zinc finger, CCHC domain containing 10 |
| chr11_+_42730639 | 0.14 |
ENSDART00000165297
|
zgc:194981
|
zgc:194981 |
| chr23_-_32092443 | 0.13 |
ENSDART00000133688
|
letmd1
|
LETM1 domain containing 1 |
| chr1_-_23370395 | 0.13 |
ENSDART00000143014
ENSDART00000126785 ENSDART00000159138 |
pds5a
|
PDS5 cohesin associated factor A |
| chr19_+_43341424 | 0.13 |
ENSDART00000134815
|
sesn2
|
sestrin 2 |
| chr20_+_23501535 | 0.13 |
ENSDART00000177922
ENSDART00000058532 |
palld
|
palladin, cytoskeletal associated protein |
| chr18_-_17075098 | 0.13 |
ENSDART00000042496
ENSDART00000192284 ENSDART00000180307 |
tango6
|
transport and golgi organization 6 homolog (Drosophila) |
| chr24_+_24170914 | 0.13 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr2_-_59247811 | 0.13 |
ENSDART00000141384
|
ftr32
|
finTRIM family, member 32 |
| chr7_+_29890292 | 0.12 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr21_+_20549395 | 0.12 |
ENSDART00000181633
|
efna5a
|
ephrin-A5a |
| chr24_-_21090447 | 0.12 |
ENSDART00000136507
ENSDART00000140786 ENSDART00000184841 |
qtrt2
|
queuine tRNA-ribosyltransferase accessory subunit 2 |
| chr23_+_1730663 | 0.12 |
ENSDART00000149545
|
tgm1
|
transglutaminase 1, K polypeptide |
| chr12_-_19862912 | 0.12 |
ENSDART00000145788
|
shisa9a
|
shisa family member 9a |
| chr3_+_27722355 | 0.12 |
ENSDART00000132761
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr1_+_1805294 | 0.11 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase Na+/K+ transporting subunit alpha 1a, tandem duplicate 3 |
| chr21_+_45268112 | 0.11 |
ENSDART00000157136
|
tcf7
|
transcription factor 7 |
| chr1_+_52690448 | 0.11 |
ENSDART00000150326
|
osbp
|
oxysterol binding protein |
| chr5_+_61361815 | 0.11 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr7_+_61184104 | 0.11 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr5_-_17876709 | 0.10 |
ENSDART00000141978
|
si:dkey-112e17.1
|
si:dkey-112e17.1 |
| chr10_+_39091716 | 0.10 |
ENSDART00000193072
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr14_+_743346 | 0.10 |
ENSDART00000110511
|
klb
|
klotho beta |
| chr2_+_44781185 | 0.10 |
ENSDART00000187103
ENSDART00000147292 ENSDART00000098123 |
ece2a
|
endothelin converting enzyme 2a |
| chr22_-_506522 | 0.10 |
ENSDART00000106645
ENSDART00000067637 |
dstyk
|
dual serine/threonine and tyrosine protein kinase |
| chr12_+_1286642 | 0.10 |
ENSDART00000157467
|
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr23_-_21446985 | 0.10 |
ENSDART00000044080
|
her12
|
hairy-related 12 |
| chr21_-_27213166 | 0.10 |
ENSDART00000146959
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr6_-_30683637 | 0.10 |
ENSDART00000065212
|
ttc4
|
tetratricopeptide repeat domain 4 |
| chr7_-_5125799 | 0.09 |
ENSDART00000173390
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr22_-_26274177 | 0.09 |
ENSDART00000060978
|
wdr83
|
WD repeat domain containing 83 |
| chr3_-_41995321 | 0.09 |
ENSDART00000192277
|
ttyh3a
|
tweety family member 3a |
| chr9_+_50175366 | 0.09 |
ENSDART00000170352
|
cobll1b
|
cordon-bleu WH2 repeat protein-like 1b |
| chr3_+_17616201 | 0.09 |
ENSDART00000156775
|
rab5c
|
RAB5C, member RAS oncogene family |
| chr2_+_57504073 | 0.09 |
ENSDART00000129811
|
onecut3b
|
one cut homeobox 3b |
| chr23_-_45705525 | 0.09 |
ENSDART00000148959
|
ednrab
|
endothelin receptor type Ab |
| chr12_-_4331117 | 0.08 |
ENSDART00000008893
|
ca15a
|
carbonic anhydrase XVa |
| chr3_+_3641429 | 0.08 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr12_+_33396489 | 0.08 |
ENSDART00000149960
|
fasn
|
fatty acid synthase |
| chr16_-_30901159 | 0.08 |
ENSDART00000180313
|
dennd3b
|
DENN/MADD domain containing 3b |
| chr1_-_53750522 | 0.08 |
ENSDART00000190755
|
akt3b
|
v-akt murine thymoma viral oncogene homolog 3b |
| chr7_-_3429874 | 0.08 |
ENSDART00000132330
|
si:ch211-285c6.1
|
si:ch211-285c6.1 |
| chr11_-_16394971 | 0.08 |
ENSDART00000180981
ENSDART00000179925 |
lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1 |
| chr20_+_5564042 | 0.08 |
ENSDART00000090934
ENSDART00000127050 |
nrxn3b
|
neurexin 3b |
| chr19_+_10536093 | 0.07 |
ENSDART00000138706
|
si:dkey-211g8.1
|
si:dkey-211g8.1 |
| chr22_-_3255341 | 0.07 |
ENSDART00000114232
|
gpr35.1
|
G protein-coupled receptor 35, tandem duplicate 1 |
| chr18_-_46241775 | 0.07 |
ENSDART00000145999
ENSDART00000134244 ENSDART00000185021 ENSDART00000181855 |
prx
|
periaxin |
| chr7_+_15324830 | 0.07 |
ENSDART00000189088
|
mespaa
|
mesoderm posterior aa |
| chr3_+_4213864 | 0.06 |
ENSDART00000059613
|
si:dkeyp-52c3.5
|
si:dkeyp-52c3.5 |
| chr12_+_29173523 | 0.06 |
ENSDART00000153212
|
adgra1a
|
adhesion G protein-coupled receptor A1a |
| chr3_+_33340939 | 0.06 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr1_-_31105376 | 0.06 |
ENSDART00000132466
|
ppp1r9alb
|
protein phosphatase 1 regulatory subunit 9A-like B |
| chr9_+_17984358 | 0.06 |
ENSDART00000192399
|
akap11
|
A kinase (PRKA) anchor protein 11 |
| chr18_-_46135927 | 0.06 |
ENSDART00000134537
ENSDART00000191481 |
SPTBN4
|
si:ch73-262h23.4 |
| chr22_+_30335936 | 0.06 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr7_-_50410524 | 0.06 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr11_+_6159595 | 0.06 |
ENSDART00000178367
|
slc1a6
|
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr12_-_30548244 | 0.06 |
ENSDART00000193616
|
zgc:158404
|
zgc:158404 |
| chr10_-_25816558 | 0.06 |
ENSDART00000017240
|
postna
|
periostin, osteoblast specific factor a |
| chr4_-_65210616 | 0.06 |
ENSDART00000183988
ENSDART00000186858 ENSDART00000185740 |
si:dkey-14o6.1
|
si:dkey-14o6.1 |
| chr11_-_23322182 | 0.05 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr25_+_20272145 | 0.05 |
ENSDART00000109605
|
si:dkey-219c3.2
|
si:dkey-219c3.2 |
| chr2_-_48539673 | 0.05 |
ENSDART00000168202
|
CR391991.4
|
|
| chr12_+_32368574 | 0.05 |
ENSDART00000086389
|
ANKFN1
|
si:ch211-277e21.2 |
| chr20_-_8110672 | 0.05 |
ENSDART00000113993
|
si:ch211-232i5.1
|
si:ch211-232i5.1 |
| chr11_+_13159988 | 0.05 |
ENSDART00000064176
|
mob3c
|
MOB kinase activator 3C |
| chr12_+_7399403 | 0.05 |
ENSDART00000103526
|
bicc1b
|
BicC family RNA binding protein 1b |
| chr7_+_3558512 | 0.05 |
ENSDART00000131884
|
si:dkey-192d15.1
|
si:dkey-192d15.1 |
| chr15_+_28116129 | 0.05 |
ENSDART00000185157
|
unc119a
|
unc-119 homolog a (C. elegans) |
| chr12_-_35054354 | 0.05 |
ENSDART00000075351
|
zgc:112285
|
zgc:112285 |
| chr10_-_39091894 | 0.05 |
ENSDART00000184505
|
igsf5a
|
immunoglobulin superfamily, member 5a |
| chr7_+_15329819 | 0.05 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr11_-_3613558 | 0.05 |
ENSDART00000163578
|
dnajc16l
|
DnaJ (Hsp40) homolog, subfamily C, member 16 like |
| chr24_-_13364311 | 0.05 |
ENSDART00000183808
|
KCNB2
|
si:dkey-192j17.1 |
| chr24_+_35933869 | 0.05 |
ENSDART00000173322
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr17_+_33433576 | 0.05 |
ENSDART00000077581
|
snap23.2
|
synaptosomal-associated protein 23.2 |
| chr22_+_9049179 | 0.05 |
ENSDART00000192625
|
si:ch211-213a13.2
|
si:ch211-213a13.2 |
| chr9_+_48415043 | 0.05 |
ENSDART00000159930
|
lrp2a
|
low density lipoprotein receptor-related protein 2a |
| chr6_+_32882821 | 0.05 |
ENSDART00000075780
|
ghrhrb
|
growth hormone releasing hormone receptor b |
| chr22_-_237651 | 0.04 |
ENSDART00000075210
|
zgc:66156
|
zgc:66156 |
| chr16_-_17300030 | 0.04 |
ENSDART00000149267
|
kel
|
Kell blood group, metallo-endopeptidase |
| chr16_+_5579744 | 0.04 |
ENSDART00000147973
|
macf1b
|
microtubule-actin crosslinking factor 1 b |
| chr3_+_2669813 | 0.04 |
ENSDART00000014205
|
CR388047.1
|
|
| chr2_+_3201345 | 0.04 |
ENSDART00000130349
|
wnt9a
|
wingless-type MMTV integration site family, member 9A |
| chr4_-_76183357 | 0.04 |
ENSDART00000181362
ENSDART00000150331 |
si:ch211-106j21.4
zgc:110171
|
si:ch211-106j21.4 zgc:110171 |
| chr24_-_37877554 | 0.04 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr7_-_1918971 | 0.04 |
ENSDART00000181759
|
CABZ01040626.1
|
|
| chr16_+_11597386 | 0.04 |
ENSDART00000173365
|
si:dkey-11o1.7
|
si:dkey-11o1.7 |
| chr14_-_2322484 | 0.04 |
ENSDART00000167806
|
si:ch73-379j16.2
|
si:ch73-379j16.2 |
| chr3_+_46315016 | 0.04 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr4_-_39484284 | 0.04 |
ENSDART00000129216
|
si:dkey-261o4.6
|
si:dkey-261o4.6 |
| chr6_-_40058686 | 0.04 |
ENSDART00000103240
|
uroc1
|
urocanate hydratase 1 |
| chr14_+_109016 | 0.04 |
ENSDART00000158405
|
gpc2
|
glypican 2 |
| chr22_+_21965148 | 0.04 |
ENSDART00000136020
|
si:ch211-207c7.2
|
si:ch211-207c7.2 |
| chr5_-_37252111 | 0.03 |
ENSDART00000185110
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr7_-_17712665 | 0.03 |
ENSDART00000149047
|
men1
|
multiple endocrine neoplasia I |
Network of associatons between targets according to the STRING database.
First level regulatory network of rbpja
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 2.8 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.3 | GO:0044650 | virion attachment to host cell(GO:0019062) adhesion of symbiont to host(GO:0044406) adhesion of symbiont to host cell(GO:0044650) |
| 0.1 | 0.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.4 | GO:0060546 | negative regulation of necroptotic process(GO:0060546) negative regulation of necrotic cell death(GO:0060547) |
| 0.1 | 0.3 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.1 | 0.3 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.6 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.1 | 0.2 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.4 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.6 | GO:0061615 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.5 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) purinergic receptor signaling pathway(GO:0035587) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.4 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.6 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.1 | GO:1903292 | protein localization to Golgi membrane(GO:1903292) |
| 0.0 | 0.8 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.5 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0002568 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.6 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.5 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.3 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.0 | 0.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:1901031 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of response to reactive oxygen species(GO:1901031) |
| 0.0 | 0.2 | GO:0031272 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.0 | 0.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0019557 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 1.2 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.0 | GO:1903538 | meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.4 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.1 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.6 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.0 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.8 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.3 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.3 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.1 | 0.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.5 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.1 | 0.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.4 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.3 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.8 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.4 | GO:0043028 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.4 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0031843 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.6 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.6 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.1 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |