Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
Results for rarga
Z-value: 0.71
Transcription factors associated with rarga
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rarga
|
ENSDARG00000034117 | retinoic acid receptor gamma a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rarga | dr11_v1_chr23_+_35847538_35847538 | -0.81 | 5.3e-05 | Click! |
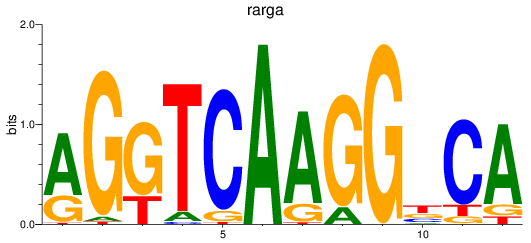
Activity profile of rarga motif
Sorted Z-values of rarga motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39786222 | 2.53 |
ENSDART00000154515
|
pimr62
|
Pim proto-oncogene, serine/threonine kinase, related 62 |
| chr16_+_25126935 | 2.12 |
ENSDART00000058945
|
zgc:92590
|
zgc:92590 |
| chr9_-_48397702 | 2.06 |
ENSDART00000147169
|
zgc:172182
|
zgc:172182 |
| chr5_+_42467867 | 1.98 |
ENSDART00000172028
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr21_+_30355767 | 1.82 |
ENSDART00000189948
|
CR749164.1
|
|
| chr7_+_20344222 | 1.61 |
ENSDART00000141186
ENSDART00000139274 |
ponzr1
|
plac8 onzin related protein 1 |
| chr25_+_3326885 | 1.37 |
ENSDART00000104866
|
ldhbb
|
lactate dehydrogenase Bb |
| chr15_-_20933574 | 1.32 |
ENSDART00000152648
ENSDART00000152448 ENSDART00000152244 |
usp2a
|
ubiquitin specific peptidase 2a |
| chr7_-_58729894 | 1.24 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr7_+_20344032 | 1.23 |
ENSDART00000144948
ENSDART00000138786 |
ponzr1
|
plac8 onzin related protein 1 |
| chr6_+_52235441 | 1.22 |
ENSDART00000056319
|
cox6c
|
cytochrome c oxidase subunit VIc |
| chr11_-_24681292 | 1.19 |
ENSDART00000089601
|
olfml3b
|
olfactomedin-like 3b |
| chr7_+_20344486 | 1.18 |
ENSDART00000134004
ENSDART00000139685 |
ponzr1
|
plac8 onzin related protein 1 |
| chr19_+_40856534 | 1.17 |
ENSDART00000051950
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr12_-_30583668 | 1.16 |
ENSDART00000153406
|
casp7
|
caspase 7, apoptosis-related cysteine peptidase |
| chr4_-_13567387 | 1.15 |
ENSDART00000132971
ENSDART00000102010 |
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr22_+_28969071 | 1.15 |
ENSDART00000163427
|
pimr95
|
Pim proto-oncogene, serine/threonine kinase, related 95 |
| chr13_-_12660318 | 1.12 |
ENSDART00000008498
|
adh8a
|
alcohol dehydrogenase 8a |
| chr17_-_26610814 | 1.10 |
ENSDART00000133402
ENSDART00000016608 |
mrpl57
|
mitochondrial ribosomal protein L57 |
| chr21_-_43117327 | 1.10 |
ENSDART00000122352
|
p4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide 2 |
| chr15_+_46386261 | 1.09 |
ENSDART00000191793
|
igsf11
|
immunoglobulin superfamily member 11 |
| chr19_-_42424599 | 1.07 |
ENSDART00000077042
|
zgc:153441
|
zgc:153441 |
| chr5_+_20693724 | 1.06 |
ENSDART00000141368
|
si:ch211-240b21.2
|
si:ch211-240b21.2 |
| chr1_+_54626491 | 1.05 |
ENSDART00000136063
|
si:ch211-202h22.9
|
si:ch211-202h22.9 |
| chr1_-_9109699 | 0.97 |
ENSDART00000147833
|
vap
|
vascular associated protein |
| chr8_-_13471916 | 0.95 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr25_+_3327071 | 0.94 |
ENSDART00000136131
ENSDART00000133243 |
ldhbb
|
lactate dehydrogenase Bb |
| chr7_-_29271738 | 0.92 |
ENSDART00000109030
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr10_+_2981090 | 0.89 |
ENSDART00000111830
|
zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr21_-_308852 | 0.87 |
ENSDART00000151613
|
lhfpl2a
|
LHFPL tetraspan subfamily member 2a |
| chr8_-_13428740 | 0.87 |
ENSDART00000131826
|
CR354547.3
|
|
| chr15_+_17321218 | 0.86 |
ENSDART00000143796
|
cltcb
|
clathrin, heavy chain b (Hc) |
| chr11_-_42315748 | 0.86 |
ENSDART00000169853
|
slmapa
|
sarcolemma associated protein a |
| chr1_+_54043563 | 0.84 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr7_-_27033080 | 0.84 |
ENSDART00000173516
|
nucb2a
|
nucleobindin 2a |
| chr22_+_29067388 | 0.80 |
ENSDART00000133673
|
pimr100
|
Pim proto-oncogene, serine/threonine kinase, related 100 |
| chr16_-_23471387 | 0.78 |
ENSDART00000180899
|
CR533578.1
|
|
| chr15_+_3284416 | 0.77 |
ENSDART00000187665
ENSDART00000171723 |
foxo1a
|
forkhead box O1 a |
| chr12_-_24928497 | 0.76 |
ENSDART00000002465
|
msh2
|
mutS homolog 2 (E. coli) |
| chr6_-_20952187 | 0.76 |
ENSDART00000074327
|
igfbp2a
|
insulin-like growth factor binding protein 2a |
| chr6_+_11990733 | 0.74 |
ENSDART00000151075
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr21_-_2299002 | 0.73 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr16_-_17258111 | 0.72 |
ENSDART00000079497
|
emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr14_-_33454595 | 0.70 |
ENSDART00000109615
ENSDART00000173267 ENSDART00000185737 ENSDART00000190989 |
tmem255a
|
transmembrane protein 255A |
| chr21_-_5879897 | 0.70 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr20_-_14665002 | 0.70 |
ENSDART00000152816
|
scrn2
|
secernin 2 |
| chr1_+_44767657 | 0.70 |
ENSDART00000073717
|
si:dkey-9i23.4
|
si:dkey-9i23.4 |
| chr3_-_15139293 | 0.70 |
ENSDART00000161213
ENSDART00000146053 |
fam173a
|
family with sequence similarity 173, member A |
| chr8_-_13419049 | 0.69 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr6_-_10835849 | 0.68 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr11_+_43363075 | 0.67 |
ENSDART00000179934
|
CU929458.1
|
|
| chr2_+_24536762 | 0.66 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr12_+_38695808 | 0.66 |
ENSDART00000188726
ENSDART00000003339 |
dnai2b
|
dynein, axonemal, intermediate chain 2b |
| chr1_-_46632948 | 0.65 |
ENSDART00000148893
ENSDART00000053232 |
cdadc1
|
cytidine and dCMP deaminase domain containing 1 |
| chr5_+_19343880 | 0.65 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr12_+_27096835 | 0.65 |
ENSDART00000149475
|
ttll6
|
tubulin tyrosine ligase-like family, member 6 |
| chr8_-_25329967 | 0.65 |
ENSDART00000139682
|
eps8l3b
|
EPS8-like 3b |
| chr8_-_13442871 | 0.64 |
ENSDART00000144887
|
pimr106
|
Pim proto-oncogene, serine/threonine kinase, related 106 |
| chr9_-_303658 | 0.64 |
ENSDART00000160338
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr7_+_15266093 | 0.62 |
ENSDART00000124676
|
sv2ba
|
synaptic vesicle glycoprotein 2Ba |
| chr3_+_62356578 | 0.61 |
ENSDART00000157030
|
iqck
|
IQ motif containing K |
| chr10_+_26813710 | 0.60 |
ENSDART00000136122
|
si:ch211-263p13.7
|
si:ch211-263p13.7 |
| chr17_-_24364527 | 0.60 |
ENSDART00000155192
|
wdpcp
|
WD repeat containing planar cell polarity effector |
| chr2_-_27774783 | 0.59 |
ENSDART00000161864
|
XKR4
|
zgc:123035 |
| chr14_-_5678457 | 0.59 |
ENSDART00000012116
|
tlx2
|
T cell leukemia homeobox 2 |
| chr3_-_18792492 | 0.59 |
ENSDART00000134208
ENSDART00000034373 |
hagh
|
hydroxyacylglutathione hydrolase |
| chr8_-_13454281 | 0.57 |
ENSDART00000141959
|
CR354547.2
|
|
| chr22_+_29009541 | 0.56 |
ENSDART00000169449
|
pimr97
|
Pim proto-oncogene, serine/threonine kinase, related 97 |
| chr21_+_44112914 | 0.56 |
ENSDART00000062836
|
fgf1b
|
fibroblast growth factor 1b |
| chr2_+_46009507 | 0.56 |
ENSDART00000193596
|
CR762395.1
|
|
| chr8_-_13486258 | 0.56 |
ENSDART00000137459
|
pimr104
|
Pim proto-oncogene, serine/threonine kinase, related 104 |
| chr19_+_40856807 | 0.56 |
ENSDART00000139083
|
gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr11_+_37201483 | 0.55 |
ENSDART00000160930
ENSDART00000173439 ENSDART00000171273 |
zgc:112265
|
zgc:112265 |
| chr3_-_62393449 | 0.55 |
ENSDART00000101870
ENSDART00000140782 ENSDART00000181704 |
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr19_-_5103313 | 0.55 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_-_18778988 | 0.55 |
ENSDART00000019235
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr17_+_23770848 | 0.54 |
ENSDART00000079646
|
kcnk18
|
potassium channel, subfamily K, member 18 |
| chr19_-_5103141 | 0.54 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr8_+_25145464 | 0.54 |
ENSDART00000136505
|
gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 3 |
| chr9_+_31795343 | 0.54 |
ENSDART00000139584
|
itgbl1
|
integrin, beta-like 1 |
| chr4_-_17409533 | 0.53 |
ENSDART00000011943
|
pah
|
phenylalanine hydroxylase |
| chr20_+_16750177 | 0.52 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr25_+_1591964 | 0.52 |
ENSDART00000093277
|
ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr9_-_22834860 | 0.52 |
ENSDART00000146486
|
neb
|
nebulin |
| chr6_-_43047774 | 0.52 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr21_+_39432248 | 0.52 |
ENSDART00000179938
|
pafah1b1b
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1b |
| chr22_-_18779232 | 0.51 |
ENSDART00000186726
|
atp5f1d
|
ATP synthase F1 subunit delta |
| chr4_+_2092683 | 0.50 |
ENSDART00000067423
|
frs2a
|
fibroblast growth factor receptor substrate 2a |
| chr17_+_26352372 | 0.49 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr1_+_57331813 | 0.49 |
ENSDART00000152440
ENSDART00000062841 |
epn3b
|
epsin 3b |
| chr8_+_20679759 | 0.49 |
ENSDART00000088668
|
nfic
|
nuclear factor I/C |
| chr7_-_56793739 | 0.47 |
ENSDART00000082842
|
si:ch211-146m13.3
|
si:ch211-146m13.3 |
| chr22_-_38258053 | 0.46 |
ENSDART00000132516
|
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr18_-_50956280 | 0.46 |
ENSDART00000058457
|
st7
|
suppression of tumorigenicity 7 |
| chr20_+_11800063 | 0.45 |
ENSDART00000152230
|
si:ch211-155o21.4
|
si:ch211-155o21.4 |
| chr20_+_6142433 | 0.44 |
ENSDART00000054084
ENSDART00000136986 |
ttr
|
transthyretin (prealbumin, amyloidosis type I) |
| chr5_-_3889047 | 0.43 |
ENSDART00000185817
|
mlxipl
|
MLX interacting protein like |
| chr18_+_14529005 | 0.42 |
ENSDART00000186379
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr2_+_38264964 | 0.41 |
ENSDART00000182068
|
dhrs1
|
dehydrogenase/reductase (SDR family) member 1 |
| chr16_-_19026414 | 0.41 |
ENSDART00000141208
|
pimr98
|
Pim proto-oncogene, serine/threonine kinase, related 98 |
| chr22_-_17595310 | 0.40 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr4_-_17629444 | 0.40 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr21_-_551014 | 0.40 |
ENSDART00000099252
|
vimr1
|
vimentin-related 1 |
| chr2_-_22230326 | 0.40 |
ENSDART00000127810
|
fam110b
|
family with sequence similarity 110, member B |
| chr10_-_4980150 | 0.39 |
ENSDART00000093228
|
mat2al
|
methionine adenosyltransferase II, alpha-like |
| chr7_-_16034324 | 0.39 |
ENSDART00000002498
ENSDART00000162962 |
elp4
|
elongator acetyltransferase complex subunit 4 |
| chr6_-_8498908 | 0.37 |
ENSDART00000149222
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr11_-_40257225 | 0.37 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
| chr5_-_41531629 | 0.36 |
ENSDART00000051082
|
akr1a1a
|
aldo-keto reductase family 1, member A1a (aldehyde reductase) |
| chr6_-_13376060 | 0.36 |
ENSDART00000104732
ENSDART00000125271 |
arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6 |
| chr14_+_46216703 | 0.36 |
ENSDART00000136045
ENSDART00000142317 |
mgst2
|
microsomal glutathione S-transferase 2 |
| chr25_+_31929325 | 0.36 |
ENSDART00000181095
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr22_+_27284462 | 0.35 |
ENSDART00000164660
|
si:ch73-103b2.1
|
si:ch73-103b2.1 |
| chr16_+_16978424 | 0.35 |
ENSDART00000143128
|
rpl18
|
ribosomal protein L18 |
| chr13_+_15182149 | 0.35 |
ENSDART00000193644
ENSDART00000134421 ENSDART00000086281 |
mavs
|
mitochondrial antiviral signaling protein |
| chr17_-_43287290 | 0.35 |
ENSDART00000156885
|
EML5
|
si:dkey-1f12.3 |
| chr11_+_37720756 | 0.35 |
ENSDART00000173337
|
snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr5_-_9090178 | 0.35 |
ENSDART00000091472
|
kcnv2b
|
potassium channel, subfamily V, member 2b |
| chr24_+_3328354 | 0.35 |
ENSDART00000147468
|
bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr22_+_28995391 | 0.34 |
ENSDART00000164486
|
pimr99
|
Pim proto-oncogene, serine/threonine kinase, related 99 |
| chr25_-_16826219 | 0.32 |
ENSDART00000191299
ENSDART00000188504 |
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr7_-_16237861 | 0.32 |
ENSDART00000173647
|
btr07
|
bloodthirsty-related gene family, member 7 |
| chr2_-_22286828 | 0.32 |
ENSDART00000168653
|
fam110b
|
family with sequence similarity 110, member B |
| chr16_-_46567344 | 0.31 |
ENSDART00000127721
|
si:dkey-152b24.7
|
si:dkey-152b24.7 |
| chr16_+_6021908 | 0.31 |
ENSDART00000163786
|
BX511115.1
|
|
| chr9_-_48281941 | 0.31 |
ENSDART00000099787
|
klhl41a
|
kelch-like family member 41a |
| chr5_-_16405651 | 0.31 |
ENSDART00000163942
|
slc39a14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr21_+_37355716 | 0.31 |
ENSDART00000131188
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr10_+_28428222 | 0.30 |
ENSDART00000135003
|
si:ch211-222e20.4
|
si:ch211-222e20.4 |
| chr8_+_48095911 | 0.30 |
ENSDART00000138717
|
si:ch211-263k4.2
|
si:ch211-263k4.2 |
| chr6_+_27418541 | 0.30 |
ENSDART00000187410
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr16_+_27345383 | 0.30 |
ENSDART00000078250
ENSDART00000162857 |
nr4a3
|
nuclear receptor subfamily 4, group A, member 3 |
| chr8_+_24740013 | 0.30 |
ENSDART00000126897
|
lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr13_-_22862133 | 0.29 |
ENSDART00000138563
|
pbld2
|
phenazine biosynthesis-like protein domain containing 2 |
| chr23_+_42336084 | 0.29 |
ENSDART00000158959
ENSDART00000161812 |
cyp2aa7
cyp2aa8
|
cytochrome P450, family 2, subfamily AA, polypeptide 7 cytochrome P450, family 2, subfamily AA, polypeptide 8 |
| chr14_+_6159162 | 0.29 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr3_+_22442445 | 0.29 |
ENSDART00000190921
|
wnk4b
|
WNK lysine deficient protein kinase 4b |
| chr25_-_19090479 | 0.28 |
ENSDART00000027465
ENSDART00000177670 |
cacna2d4b
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4b |
| chr5_+_69868911 | 0.28 |
ENSDART00000014649
ENSDART00000188215 ENSDART00000167385 |
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr22_-_24967348 | 0.28 |
ENSDART00000153490
ENSDART00000084871 |
fam20cl
|
family with sequence similarity 20, member C like |
| chr19_+_30387999 | 0.28 |
ENSDART00000145396
|
tspan13b
|
tetraspanin 13b |
| chr15_-_31514818 | 0.27 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr4_+_8680767 | 0.27 |
ENSDART00000182726
|
adipor2
|
adiponectin receptor 2 |
| chr7_+_26100024 | 0.27 |
ENSDART00000173726
|
si:ch211-196f2.3
|
si:ch211-196f2.3 |
| chr15_+_1004680 | 0.27 |
ENSDART00000157310
|
si:dkey-77f5.8
|
si:dkey-77f5.8 |
| chr11_+_37720552 | 0.27 |
ENSDART00000050296
ENSDART00000172867 |
snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr5_+_1079423 | 0.27 |
ENSDART00000172231
|
si:zfos-128g4.2
|
si:zfos-128g4.2 |
| chr10_-_35220285 | 0.27 |
ENSDART00000180439
|
ypel2a
|
yippee-like 2a |
| chr4_-_23839789 | 0.27 |
ENSDART00000143571
|
usp6nl
|
USP6 N-terminal like |
| chr12_+_28799988 | 0.26 |
ENSDART00000022724
|
pnpo
|
pyridoxamine 5'-phosphate oxidase |
| chr18_+_17959681 | 0.26 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr21_-_22831388 | 0.26 |
ENSDART00000151040
|
angptl5
|
angiopoietin-like 5 |
| chr15_+_24644251 | 0.26 |
ENSDART00000181660
|
smtnl
|
smoothelin, like |
| chr2_-_31634978 | 0.26 |
ENSDART00000135668
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr2_+_27010439 | 0.26 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr15_-_30815826 | 0.25 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr5_+_37290260 | 0.25 |
ENSDART00000113642
|
zgc:163098
|
zgc:163098 |
| chr3_+_29458517 | 0.24 |
ENSDART00000134258
|
grap2a
|
GRB2-related adaptor protein 2a |
| chr12_+_4573696 | 0.24 |
ENSDART00000152534
|
si:dkey-94f20.4
|
si:dkey-94f20.4 |
| chr3_-_15080226 | 0.24 |
ENSDART00000109818
ENSDART00000139835 |
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr20_+_26095530 | 0.23 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr20_+_36682051 | 0.23 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr18_+_13315739 | 0.23 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr14_-_25111496 | 0.23 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| chr12_-_7806007 | 0.23 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr7_+_48806420 | 0.22 |
ENSDART00000083431
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr13_+_22480496 | 0.22 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr2_+_38608290 | 0.22 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr6_+_18569453 | 0.22 |
ENSDART00000171338
|
rhot1b
|
ras homolog family member T1 |
| chr13_+_22480857 | 0.22 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr25_-_29415369 | 0.22 |
ENSDART00000110774
ENSDART00000019183 |
ugt5a2
ugt5a1
|
UDP glucuronosyltransferase 5 family, polypeptide A2 UDP glucuronosyltransferase 5 family, polypeptide A1 |
| chr21_+_5932140 | 0.21 |
ENSDART00000193767
|
rexo4
|
REX4 homolog, 3'-5' exonuclease |
| chr21_-_2958422 | 0.21 |
ENSDART00000174091
|
zgc:194215
|
zgc:194215 |
| chr19_-_14155781 | 0.19 |
ENSDART00000169232
|
nr0b2b
|
nuclear receptor subfamily 0, group B, member 2b |
| chr15_+_44133972 | 0.19 |
ENSDART00000157846
ENSDART00000191082 |
CU655961.2
|
|
| chr3_+_155659 | 0.19 |
ENSDART00000183570
|
FP326649.2
|
|
| chr7_-_41338923 | 0.19 |
ENSDART00000099138
|
ncf2
|
neutrophil cytosolic factor 2 |
| chr9_-_27749936 | 0.19 |
ENSDART00000064156
|
tbccd1
|
TBCC domain containing 1 |
| chr6_+_52804267 | 0.19 |
ENSDART00000065681
|
matn4
|
matrilin 4 |
| chr17_-_12196865 | 0.18 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
| chr21_+_32332616 | 0.18 |
ENSDART00000154105
|
si:ch211-247j9.1
|
si:ch211-247j9.1 |
| chr6_-_8498676 | 0.18 |
ENSDART00000148627
|
pglyrp2
|
peptidoglycan recognition protein 2 |
| chr25_-_1687481 | 0.18 |
ENSDART00000163553
ENSDART00000179460 |
mon2
|
MON2 homolog, regulator of endosome-to-Golgi trafficking |
| chr17_-_51224800 | 0.18 |
ENSDART00000150089
|
psen1
|
presenilin 1 |
| chr16_-_17116499 | 0.18 |
ENSDART00000138983
|
si:dkey-260g12.1
|
si:dkey-260g12.1 |
| chr2_-_47431205 | 0.17 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr22_+_10163901 | 0.17 |
ENSDART00000190468
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr20_+_6543674 | 0.17 |
ENSDART00000134204
|
tns3.1
|
tensin 3, tandem duplicate 1 |
| chr7_-_35708450 | 0.17 |
ENSDART00000193886
|
irx5a
|
iroquois homeobox 5a |
| chr12_-_29301022 | 0.17 |
ENSDART00000187826
|
sh2d4bb
|
SH2 domain containing 4Bb |
| chr21_-_20939488 | 0.16 |
ENSDART00000039043
|
rgs7bpb
|
regulator of G protein signaling 7 binding protein b |
| chr12_+_46708920 | 0.16 |
ENSDART00000153089
|
exoc7
|
exocyst complex component 7 |
| chr21_+_28724099 | 0.16 |
ENSDART00000138017
|
puraa
|
purine-rich element binding protein Aa |
| chr21_-_29100110 | 0.16 |
ENSDART00000142598
|
timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr19_-_8732037 | 0.15 |
ENSDART00000138971
|
si:ch211-39a7.1
|
si:ch211-39a7.1 |
| chr12_+_3912544 | 0.15 |
ENSDART00000013465
|
tbx6
|
T-box 6 |
| chr21_+_5801105 | 0.15 |
ENSDART00000151225
ENSDART00000184487 |
ccng2
|
cyclin G2 |
| chr13_-_15994419 | 0.15 |
ENSDART00000079724
ENSDART00000042377 ENSDART00000046079 ENSDART00000050481 ENSDART00000016430 |
ikzf1
|
IKAROS family zinc finger 1 (Ikaros) |
| chr3_+_41922114 | 0.14 |
ENSDART00000138280
|
lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr16_-_40727455 | 0.13 |
ENSDART00000162331
|
si:dkey-22o22.2
|
si:dkey-22o22.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of rarga
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.0 | GO:0061333 | renal tubule morphogenesis(GO:0061333) nephron tubule morphogenesis(GO:0072078) nephron epithelium morphogenesis(GO:0072088) |
| 0.3 | 1.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 0.6 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.2 | 0.6 | GO:0098543 | detection of biotic stimulus(GO:0009595) detection of bacterium(GO:0016045) detection of other organism(GO:0098543) detection of external biotic stimulus(GO:0098581) |
| 0.2 | 1.1 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.2 | 0.6 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.2 | 1.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.2 | 0.8 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.8 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.9 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 0.6 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.1 | 0.3 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.1 | 0.3 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.3 | GO:0042822 | water-soluble vitamin biosynthetic process(GO:0042364) pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.1 | 0.5 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.1 | 0.8 | GO:0032096 | negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 | 0.7 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.1 | 0.8 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.6 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 0.4 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.1 | 0.7 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.1 | 0.4 | GO:0039528 | cytoplasmic pattern recognition receptor signaling pathway in response to virus(GO:0039528) cellular response to virus(GO:0098586) |
| 0.1 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.1 | 0.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.6 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.1 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.1 | 1.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.7 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.2 | GO:0051661 | Golgi localization(GO:0051645) maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.5 | GO:1902222 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.2 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 1.0 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.7 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.5 | GO:0030324 | respiratory tube development(GO:0030323) lung development(GO:0030324) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 0.1 | GO:0002369 | T cell cytokine production(GO:0002369) |
| 0.0 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.3 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.2 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.0 | 10.7 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0090386 | phagosome maturation involved in apoptotic cell clearance(GO:0090386) phagolysosome assembly involved in apoptotic cell clearance(GO:0090387) |
| 0.0 | 0.3 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.2 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.7 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.2 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.5 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0071267 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine biosynthetic process(GO:0071265) L-methionine salvage(GO:0071267) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.2 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.3 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.3 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.2 | GO:0072498 | embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.5 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) establishment of mitochondrion localization(GO:0051654) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.2 | 0.9 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.2 | 0.6 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 0.6 | GO:0097541 | axonemal basal plate(GO:0097541) |
| 0.2 | 0.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.1 | 0.5 | GO:0008247 | 1-alkyl-2-acetylglycerophosphocholine esterase complex(GO:0008247) |
| 0.1 | 1.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 0.7 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.7 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 1.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 1.1 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.3 | 0.8 | GO:0032404 | guanine/thymine mispair binding(GO:0032137) single base insertion or deletion binding(GO:0032138) single thymine insertion binding(GO:0032143) oxidized DNA binding(GO:0032356) oxidized purine DNA binding(GO:0032357) mismatch repair complex binding(GO:0032404) MutLalpha complex binding(GO:0032405) |
| 0.2 | 1.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) S-(hydroxymethyl)glutathione dehydrogenase activity(GO:0051903) |
| 0.2 | 0.6 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.6 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 0.7 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.5 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.1 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.3 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.1 | 1.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.9 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.4 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.4 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 1.2 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 0.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.8 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.9 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.7 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.6 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 1.2 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.7 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor binding(GO:0048403) brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.0 | 0.6 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.3 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 2.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 10.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 1.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.4 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.5 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |