Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
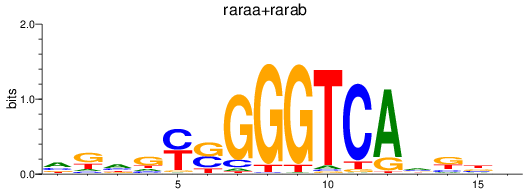
Results for raraa+rarab
Z-value: 0.78
Transcription factors associated with raraa+rarab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rarab
|
ENSDARG00000034893 | retinoic acid receptor, alpha b |
|
raraa
|
ENSDARG00000056783 | retinoic acid receptor, alpha a |
|
rarab
|
ENSDARG00000111757 | retinoic acid receptor, alpha b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rarab | dr11_v1_chr3_-_33175583_33175583 | -0.99 | 7.8e-14 | Click! |
| raraa | dr11_v1_chr12_+_10952794_10952794 | -0.81 | 5.2e-05 | Click! |
Activity profile of raraa+rarab motif
Sorted Z-values of raraa+rarab motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_13471916 | 2.07 |
ENSDART00000146558
|
pimr105
|
Pim proto-oncogene, serine/threonine kinase, related 105 |
| chr8_-_13428740 | 1.94 |
ENSDART00000131826
|
CR354547.3
|
|
| chr18_+_19842569 | 1.91 |
ENSDART00000147470
|
iqch
|
IQ motif containing H |
| chr18_+_1145571 | 1.90 |
ENSDART00000135055
|
rec114
|
REC114 meiotic recombination protein |
| chr14_-_36862745 | 1.64 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr8_-_13442871 | 1.52 |
ENSDART00000144887
|
pimr106
|
Pim proto-oncogene, serine/threonine kinase, related 106 |
| chr8_-_13419049 | 1.50 |
ENSDART00000133656
|
pimr101
|
Pim proto-oncogene, serine/threonine kinase, related 101 |
| chr4_-_191736 | 1.49 |
ENSDART00000169187
ENSDART00000192054 |
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr21_+_28445052 | 1.48 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr22_-_31788170 | 1.44 |
ENSDART00000170925
|
pimr207
|
Pim proto-oncogene, serine/threonine kinase, related 207 |
| chr8_-_13486258 | 1.35 |
ENSDART00000137459
|
pimr104
|
Pim proto-oncogene, serine/threonine kinase, related 104 |
| chr17_+_46739693 | 1.27 |
ENSDART00000097810
|
pimr22
|
Pim proto-oncogene, serine/threonine kinase, related 22 |
| chr5_-_3885727 | 1.26 |
ENSDART00000143250
|
mlxipl
|
MLX interacting protein like |
| chr8_-_13454281 | 1.26 |
ENSDART00000141959
|
CR354547.2
|
|
| chr17_+_53311618 | 1.04 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr22_+_38173960 | 0.94 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr24_+_42149453 | 0.89 |
ENSDART00000128766
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr21_-_32284532 | 0.82 |
ENSDART00000190676
|
clk4b
|
CDC-like kinase 4b |
| chr24_+_21973929 | 0.82 |
ENSDART00000042495
|
sat1b
|
spermidine/spermine N1-acetyltransferase 1b |
| chr8_-_28342313 | 0.81 |
ENSDART00000005583
|
kdm5bb
|
lysine (K)-specific demethylase 5Bb |
| chr2_+_52847049 | 0.78 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr23_+_25822742 | 0.77 |
ENSDART00000184436
|
r3hdml
|
R3H domain containing-like |
| chr11_+_1608348 | 0.74 |
ENSDART00000162438
|
si:dkey-40c23.3
|
si:dkey-40c23.3 |
| chr17_+_14886828 | 0.74 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr22_-_834106 | 0.72 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr23_+_25305431 | 0.68 |
ENSDART00000143291
|
RPL41
|
si:dkey-151g10.6 |
| chr18_-_36066087 | 0.66 |
ENSDART00000059352
ENSDART00000145177 |
exosc5
|
exosome component 5 |
| chr5_+_64856666 | 0.65 |
ENSDART00000050863
|
zgc:101858
|
zgc:101858 |
| chr22_+_30009926 | 0.64 |
ENSDART00000142529
|
si:dkey-286j15.1
|
si:dkey-286j15.1 |
| chr12_+_2677303 | 0.62 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr18_+_3338228 | 0.62 |
ENSDART00000161520
|
gdpd4a
|
glycerophosphodiester phosphodiesterase domain containing 4a |
| chr24_+_42149296 | 0.61 |
ENSDART00000154231
|
serpinb1l3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 3 |
| chr17_+_53311243 | 0.58 |
ENSDART00000160241
ENSDART00000160009 ENSDART00000162239 |
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr13_+_24834199 | 0.57 |
ENSDART00000101274
|
zgc:153981
|
zgc:153981 |
| chr6_-_8277221 | 0.51 |
ENSDART00000053869
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr1_+_59154521 | 0.47 |
ENSDART00000130089
ENSDART00000152456 |
soul5l
|
heme-binding protein soul5, like |
| chr14_+_94946 | 0.44 |
ENSDART00000165766
ENSDART00000163778 |
mcm7
|
minichromosome maintenance complex component 7 |
| chr2_+_56213694 | 0.44 |
ENSDART00000162582
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr19_+_5291935 | 0.37 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr2_+_3696038 | 0.33 |
ENSDART00000150499
ENSDART00000136906 ENSDART00000164152 ENSDART00000128514 ENSDART00000108964 |
egfra
|
epidermal growth factor receptor a (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) |
| chr18_+_10820430 | 0.31 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr16_+_7242610 | 0.31 |
ENSDART00000081477
|
sri
|
sorcin |
| chr14_-_51855047 | 0.30 |
ENSDART00000088912
|
cplx1
|
complexin 1 |
| chr23_-_3703569 | 0.27 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr24_+_37723362 | 0.26 |
ENSDART00000136836
|
rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr17_+_33340675 | 0.26 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr11_+_25111846 | 0.26 |
ENSDART00000128705
ENSDART00000190058 |
ndrg3a
|
ndrg family member 3a |
| chr3_+_5297493 | 0.25 |
ENSDART00000138596
|
si:ch211-150d5.3
|
si:ch211-150d5.3 |
| chr19_+_19762183 | 0.22 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr22_+_20208185 | 0.21 |
ENSDART00000142748
|
si:dkey-110c1.7
|
si:dkey-110c1.7 |
| chr18_+_8340886 | 0.20 |
ENSDART00000081132
|
cpt1b
|
carnitine palmitoyltransferase 1B (muscle) |
| chr14_-_2001386 | 0.18 |
ENSDART00000170346
ENSDART00000163975 |
pcdh2g5
|
protocadherin 2 gamma 5 |
| chr25_-_3132060 | 0.18 |
ENSDART00000014363
|
epx
|
eosinophil peroxidase |
| chr2_+_58841181 | 0.14 |
ENSDART00000164102
|
cirbpa
|
cold inducible RNA binding protein a |
| chr1_+_59538755 | 0.12 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr17_+_14711765 | 0.10 |
ENSDART00000012889
|
cx28.6
|
connexin 28.6 |
| chr22_+_21324398 | 0.10 |
ENSDART00000168509
|
shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr3_+_57268363 | 0.10 |
ENSDART00000180725
|
TMEM235 (1 of many)
|
transmembrane protein 235 |
| chr22_-_9900378 | 0.10 |
ENSDART00000193906
|
znf991
|
zinc finger protein 991 |
| chr4_+_1530287 | 0.09 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr10_-_41450367 | 0.08 |
ENSDART00000122682
ENSDART00000189549 |
cabp1b
|
calcium binding protein 1b |
| chr14_+_41518257 | 0.07 |
ENSDART00000050037
|
chrnb3b
|
cholinergic receptor, nicotinic, beta 3b (neuronal) |
| chr14_+_94603 | 0.06 |
ENSDART00000162480
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr25_+_1549838 | 0.06 |
ENSDART00000115001
|
avpr1aa
|
arginine vasopressin receptor 1Aa |
| chr24_+_42117231 | 0.04 |
ENSDART00000067734
|
snx16
|
sorting nexin 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of raraa+rarab
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0052576 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.2 | 0.8 | GO:0009447 | polyamine catabolic process(GO:0006598) putrescine catabolic process(GO:0009447) |
| 0.1 | 0.7 | GO:0034695 | response to prostaglandin E(GO:0034695) cellular response to prostaglandin E stimulus(GO:0071380) |
| 0.1 | 0.5 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 0.7 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.6 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 1.5 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.2 | GO:0016081 | synaptic vesicle docking(GO:0016081) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 9.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.3 | GO:0072554 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.0 | 0.8 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.8 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.8 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.5 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 0.7 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.8 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 1.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 2.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.9 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |