Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
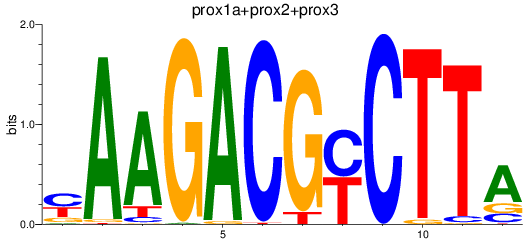
Results for prox1a+prox2+prox3
Z-value: 0.60
Transcription factors associated with prox1a+prox2+prox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prox2
|
ENSDARG00000041952 | prospero homeobox 2 |
|
prox1a
|
ENSDARG00000055158 | prospero homeobox 1a |
|
prox3
|
ENSDARG00000088810 | prospero homeobox 3 |
|
prox3
|
ENSDARG00000113014 | prospero homeobox 3 |
|
prox2
|
ENSDARG00000117137 | prospero homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prox2 | dr11_v1_chr17_-_52521002_52521002 | -0.79 | 8.3e-05 | Click! |
| prox1a | dr11_v1_chr17_-_32863250_32863250 | -0.75 | 3.0e-04 | Click! |
| prox1b | dr11_v1_chr7_-_19940473_19940473 | 0.71 | 9.5e-04 | Click! |
Activity profile of prox1a+prox2+prox3 motif
Sorted Z-values of prox1a+prox2+prox3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_54919260 | 2.09 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr8_+_47219107 | 1.69 |
ENSDART00000146018
ENSDART00000075068 |
mthfr
|
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr2_+_25658112 | 1.60 |
ENSDART00000051234
|
tnika
|
TRAF2 and NCK interacting kinase a |
| chr14_+_24840669 | 1.50 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr1_-_9486214 | 1.48 |
ENSDART00000137821
|
micall2b
|
mical-like 2b |
| chr7_-_44605050 | 1.47 |
ENSDART00000148471
ENSDART00000149072 |
tk2
|
thymidine kinase 2, mitochondrial |
| chr1_-_9485939 | 1.47 |
ENSDART00000157814
|
micall2b
|
mical-like 2b |
| chr3_-_26191960 | 1.35 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr16_+_39196727 | 1.33 |
ENSDART00000017017
|
zdhhc3a
|
zinc finger, DHHC-type containing 3a |
| chr14_+_28473173 | 1.33 |
ENSDART00000017075
|
xiap
|
X-linked inhibitor of apoptosis |
| chr19_+_42886413 | 1.26 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr7_+_6941583 | 1.12 |
ENSDART00000160709
ENSDART00000157634 |
rbm14b
|
RNA binding motif protein 14b |
| chr12_-_23365737 | 0.95 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr19_-_31042570 | 0.94 |
ENSDART00000144337
ENSDART00000136213 ENSDART00000133101 ENSDART00000190949 |
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr18_+_16750080 | 0.94 |
ENSDART00000136320
|
rnf141
|
ring finger protein 141 |
| chr17_+_15674052 | 0.93 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr11_-_25461336 | 0.90 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr15_+_2190229 | 0.88 |
ENSDART00000147710
|
rsrc1
|
arginine/serine-rich coiled-coil 1 |
| chr13_-_35765028 | 0.87 |
ENSDART00000157391
|
erlec1
|
endoplasmic reticulum lectin 1 |
| chr10_+_7719796 | 0.86 |
ENSDART00000191795
|
ggcx
|
gamma-glutamyl carboxylase |
| chr13_-_12667220 | 0.86 |
ENSDART00000079594
|
fam241a
|
family with sequence similarity 241 member A |
| chr13_+_11829072 | 0.78 |
ENSDART00000079356
ENSDART00000170160 |
sufu
|
suppressor of fused homolog (Drosophila) |
| chr4_+_5196469 | 0.77 |
ENSDART00000067386
|
rad51ap1
|
RAD51 associated protein 1 |
| chr15_-_31177324 | 0.76 |
ENSDART00000008854
|
wsb1
|
WD repeat and SOCS box containing 1 |
| chr9_+_8968702 | 0.76 |
ENSDART00000008490
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr9_+_21401189 | 0.73 |
ENSDART00000062669
|
cx30.3
|
connexin 30.3 |
| chr2_+_50722439 | 0.72 |
ENSDART00000188927
|
fyco1b
|
FYVE and coiled-coil domain containing 1b |
| chr14_-_6286966 | 0.71 |
ENSDART00000168174
|
elp1
|
elongator complex protein 1 |
| chr6_+_59642695 | 0.66 |
ENSDART00000166373
ENSDART00000161030 |
R3HDM2
|
R3H domain containing 2 |
| chr7_-_44604540 | 0.65 |
ENSDART00000149186
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr7_-_44604821 | 0.64 |
ENSDART00000148967
|
tk2
|
thymidine kinase 2, mitochondrial |
| chr8_+_11325310 | 0.64 |
ENSDART00000142577
|
fxn
|
frataxin |
| chr14_-_30971264 | 0.58 |
ENSDART00000010512
|
zgc:92907
|
zgc:92907 |
| chr10_-_38316134 | 0.58 |
ENSDART00000149580
|
nrip1b
|
nuclear receptor interacting protein 1b |
| chr6_-_32411703 | 0.56 |
ENSDART00000151002
ENSDART00000078908 |
usp1
|
ubiquitin specific peptidase 1 |
| chr20_+_38201644 | 0.54 |
ENSDART00000022694
|
ehd3
|
EH-domain containing 3 |
| chr24_-_18809433 | 0.54 |
ENSDART00000152009
|
arfgef1
|
ADP-ribosylation factor guanine nucleotide-exchange factor 1 (brefeldin A-inhibited) |
| chr16_-_13680692 | 0.51 |
ENSDART00000047452
|
ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr23_+_45845423 | 0.50 |
ENSDART00000183404
|
lmnl3
|
lamin L3 |
| chr7_+_20917966 | 0.49 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr3_+_22335030 | 0.47 |
ENSDART00000055676
|
zgc:103564
|
zgc:103564 |
| chr10_+_43037064 | 0.46 |
ENSDART00000160159
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr12_-_4408828 | 0.43 |
ENSDART00000152447
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr7_+_7019911 | 0.38 |
ENSDART00000172421
|
rbm14b
|
RNA binding motif protein 14b |
| chr22_+_10678141 | 0.38 |
ENSDART00000193341
|
hyal2b
|
hyaluronoglucosaminidase 2b |
| chr19_-_27395531 | 0.37 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr3_-_15667713 | 0.36 |
ENSDART00000026658
|
zgc:66474
|
zgc:66474 |
| chr15_-_23475051 | 0.35 |
ENSDART00000152460
|
nlrx1
|
NLR family member X1 |
| chr13_+_35765317 | 0.35 |
ENSDART00000100156
ENSDART00000167650 |
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr18_-_34549721 | 0.33 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr12_-_31422433 | 0.32 |
ENSDART00000186075
ENSDART00000153172 ENSDART00000066256 |
vti1a
|
vesicle transport through interaction with t-SNAREs 1A |
| chrM_+_8894 | 0.32 |
ENSDART00000093611
|
mt-atp8
|
ATP synthase 8, mitochondrial |
| chr8_+_30600386 | 0.31 |
ENSDART00000164976
|
specc1la
|
sperm antigen with calponin homology and coiled-coil domains 1-like a |
| chr2_+_6885852 | 0.28 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr17_-_37195163 | 0.27 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr4_+_3455146 | 0.26 |
ENSDART00000180124
|
znf800b
|
zinc finger protein 800b |
| chr13_-_27384697 | 0.26 |
ENSDART00000146230
|
kcnq5a
|
potassium voltage-gated channel, KQT-like subfamily, member 5a |
| chr17_-_37195354 | 0.25 |
ENSDART00000190963
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr23_-_13877898 | 0.24 |
ENSDART00000138696
|
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr25_+_22587306 | 0.23 |
ENSDART00000067479
|
stra6
|
stimulated by retinoic acid 6 |
| chr19_-_13950231 | 0.23 |
ENSDART00000168665
|
sh3bgrl3
|
SH3 domain binding glutamate-rich protein like 3 |
| chr25_+_16312258 | 0.23 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr20_-_54014373 | 0.22 |
ENSDART00000152934
|
si:dkey-241l7.6
|
si:dkey-241l7.6 |
| chr22_+_17286841 | 0.18 |
ENSDART00000133176
|
FAM163A
|
si:dkey-171o17.8 |
| chr12_+_6065661 | 0.17 |
ENSDART00000142418
|
sgms1
|
sphingomyelin synthase 1 |
| chr3_+_21200763 | 0.17 |
ENSDART00000067841
|
zgc:112038
|
zgc:112038 |
| chr5_-_69707787 | 0.16 |
ENSDART00000108820
ENSDART00000149692 |
dguok
|
deoxyguanosine kinase |
| chr4_+_3455665 | 0.15 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr25_-_10630496 | 0.13 |
ENSDART00000153639
ENSDART00000181722 ENSDART00000177834 |
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr17_-_21832119 | 0.13 |
ENSDART00000153539
ENSDART00000079008 |
si:ch211-208g24.8
|
si:ch211-208g24.8 |
| chr25_+_8407892 | 0.11 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr16_-_31000727 | 0.11 |
ENSDART00000077185
|
dgat1b
|
diacylglycerol O-acyltransferase 1b |
| chr22_-_23591493 | 0.10 |
ENSDART00000170266
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr19_+_7929704 | 0.10 |
ENSDART00000147015
|
si:dkey-266f7.4
|
si:dkey-266f7.4 |
| chr21_+_21791343 | 0.09 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr12_+_13404784 | 0.08 |
ENSDART00000167977
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr17_-_10059557 | 0.08 |
ENSDART00000092209
ENSDART00000161243 |
baz1a
|
bromodomain adjacent to zinc finger domain, 1A |
| chr5_+_37517800 | 0.08 |
ENSDART00000048107
|
FP102018.1
|
Danio rerio latent transforming growth factor beta binding protein 3 (ltbp3), mRNA. |
| chr10_-_20637098 | 0.07 |
ENSDART00000080391
|
sprn2
|
shadow of prion protein 2 |
| chrM_+_9052 | 0.07 |
ENSDART00000093612
|
mt-atp6
|
ATP synthase 6, mitochondrial |
| chr25_+_19947096 | 0.07 |
ENSDART00000154221
|
kcna6a
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 a |
| chr5_-_61638125 | 0.06 |
ENSDART00000134314
|
si:dkey-261j4.3
|
si:dkey-261j4.3 |
| chr11_+_6739433 | 0.06 |
ENSDART00000163397
|
pde4cb
|
phosphodiesterase 4C, cAMP-specific b |
| chr7_-_16205471 | 0.06 |
ENSDART00000173584
|
btr05
|
bloodthirsty-related gene family, member 5 |
| chr14_+_31751260 | 0.05 |
ENSDART00000169796
|
si:dkeyp-11e3.1
|
si:dkeyp-11e3.1 |
| chr8_-_4475908 | 0.05 |
ENSDART00000191027
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr22_-_23591340 | 0.04 |
ENSDART00000167024
|
f13b
|
coagulation factor XIII, B polypeptide |
| chr19_-_22621991 | 0.03 |
ENSDART00000175106
|
pleca
|
plectin a |
| chr17_+_5895500 | 0.03 |
ENSDART00000019905
|
fndc4b
|
fibronectin type III domain containing 4b |
| chr19_-_22621811 | 0.03 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr15_-_5245726 | 0.02 |
ENSDART00000174079
ENSDART00000174041 |
or128-1
|
odorant receptor, family E, subfamily 128, member 1 |
| chr4_-_71551644 | 0.02 |
ENSDART00000168064
|
si:dkey-27n6.1
|
si:dkey-27n6.1 |
| chr3_+_21669545 | 0.01 |
ENSDART00000156527
|
crhr1
|
corticotropin releasing hormone receptor 1 |
| chr2_+_927204 | 0.01 |
ENSDART00000165477
|
AL935300.3
|
|
| chr4_-_75899294 | 0.01 |
ENSDART00000157887
|
si:dkey-261j11.3
|
si:dkey-261j11.3 |
| chr25_+_19947298 | 0.01 |
ENSDART00000067648
|
kcna6a
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 a |
| chr18_-_44611252 | 0.00 |
ENSDART00000173095
|
spred3
|
sprouty-related, EVH1 domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of prox1a+prox2+prox3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 0.7 | GO:0002926 | tRNA wobble base 5-methoxycarbonylmethyl-2-thiouridine biosynthesis.(GO:0002926) |
| 0.2 | 0.8 | GO:1900181 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
| 0.1 | 0.7 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 0.9 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 1.7 | GO:0035999 | methionine biosynthetic process(GO:0009086) tetrahydrofolate interconversion(GO:0035999) |
| 0.1 | 0.5 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.1 | 0.5 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 0.6 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 3.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.9 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 1.3 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.0 | 1.3 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.6 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.4 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.0 | 0.8 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.8 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 1.6 | GO:0032147 | activation of protein kinase activity(GO:0032147) |
| 0.0 | 0.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0032456 | endocytic recycling(GO:0032456) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.2 | 0.8 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.1 | 0.7 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.9 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.9 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.3 | GO:0019107 | myristoyltransferase activity(GO:0019107) |
| 0.2 | 0.9 | GO:0008488 | gamma-glutamyl carboxylase activity(GO:0008488) |
| 0.2 | 0.5 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.1 | 0.5 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.1 | 0.4 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.1 | 0.4 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.6 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.1 | 0.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 1.7 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 1.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.8 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.9 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.1 | 1.3 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 0.6 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 1.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.5 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |