Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
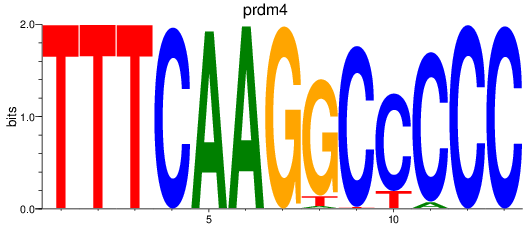
Results for prdm4
Z-value: 0.21
Transcription factors associated with prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm4
|
ENSDARG00000017366 | PR domain containing 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm4 | dr11_v1_chr4_-_14926637_14926637 | 0.91 | 1.1e-07 | Click! |
Activity profile of prdm4 motif
Sorted Z-values of prdm4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_24682244 | 0.63 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr3_+_31621774 | 0.43 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chrM_+_9735 | 0.41 |
ENSDART00000093613
|
mt-co3
|
cytochrome c oxidase III, mitochondrial |
| chr10_-_3332362 | 0.31 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr23_-_31400792 | 0.24 |
ENSDART00000132736
|
lca5
|
Leber congenital amaurosis 5 |
| chr8_+_20679759 | 0.20 |
ENSDART00000088668
|
nfic
|
nuclear factor I/C |
| chr9_+_22359919 | 0.19 |
ENSDART00000009591
|
crygs4
|
crystallin, gamma S4 |
| chr3_+_32425202 | 0.19 |
ENSDART00000156464
|
prr12b
|
proline rich 12b |
| chr10_-_9115383 | 0.12 |
ENSDART00000139324
|
si:dkeyp-41f9.3
|
si:dkeyp-41f9.3 |
| chr4_-_2868112 | 0.12 |
ENSDART00000133843
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr17_+_31611854 | 0.11 |
ENSDART00000011706
|
adprhl2
|
ADP-ribosylhydrolase like 2 |
| chr9_-_13355071 | 0.10 |
ENSDART00000084055
|
fzd7a
|
frizzled class receptor 7a |
| chr20_-_45709990 | 0.09 |
ENSDART00000027482
|
gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr4_-_2867461 | 0.08 |
ENSDART00000160308
|
pde3a
|
phosphodiesterase 3A, cGMP-inhibited |
| chr17_-_31611692 | 0.06 |
ENSDART00000141480
|
si:dkey-170l10.1
|
si:dkey-170l10.1 |
| chr22_-_21046843 | 0.05 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr22_-_21046654 | 0.05 |
ENSDART00000064902
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr19_+_2670130 | 0.03 |
ENSDART00000152152
|
si:ch73-24k9.2
|
si:ch73-24k9.2 |
| chr9_+_15893093 | 0.01 |
ENSDART00000099483
ENSDART00000134657 |
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr20_-_26001288 | 0.00 |
ENSDART00000136518
ENSDART00000063177 |
capn3b
|
calpain 3b |
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0061341 | non-canonical Wnt signaling pathway involved in heart development(GO:0061341) |
| 0.0 | 0.2 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |