Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
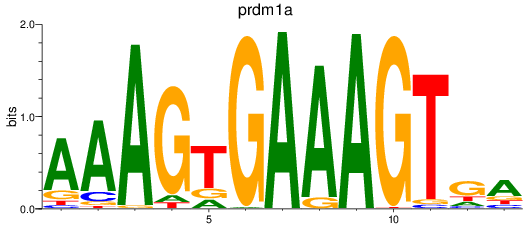
Results for prdm1a
Z-value: 0.45
Transcription factors associated with prdm1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm1a
|
ENSDARG00000002445 | PR domain containing 1a, with ZNF domain |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm1a | dr11_v1_chr16_-_7443388_7443388 | 0.95 | 3.3e-09 | Click! |
Activity profile of prdm1a motif
Sorted Z-values of prdm1a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr24_+_24923166 | 1.38 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr8_-_50259448 | 1.29 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr10_-_29816467 | 1.19 |
ENSDART00000055913
|
hist2h2l
|
histone 2, H2, like |
| chr14_+_45566074 | 1.16 |
ENSDART00000133477
ENSDART00000185899 |
zgc:92249
|
zgc:92249 |
| chr16_+_42829735 | 1.15 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr6_+_24661736 | 1.14 |
ENSDART00000169283
|
znf644b
|
zinc finger protein 644b |
| chr4_+_686157 | 1.08 |
ENSDART00000169748
|
fbxo7
|
F-box protein 7 |
| chr24_-_39610585 | 1.06 |
ENSDART00000066506
|
cox6b1
|
cytochrome c oxidase subunit VIb polypeptide 1 |
| chr10_-_34981219 | 0.97 |
ENSDART00000132823
|
smad9
|
SMAD family member 9 |
| chr10_-_34981028 | 0.97 |
ENSDART00000186108
|
smad9
|
SMAD family member 9 |
| chr15_-_47929455 | 0.96 |
ENSDART00000064462
|
psma6l
|
proteasome subunit alpha 6, like |
| chr25_+_245438 | 0.93 |
ENSDART00000004689
|
zgc:92481
|
zgc:92481 |
| chr16_+_42830152 | 0.93 |
ENSDART00000159730
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr7_-_28148310 | 0.93 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr24_+_12945803 | 0.91 |
ENSDART00000005105
|
psme1
|
proteasome activator subunit 1 |
| chr7_+_49695904 | 0.87 |
ENSDART00000183550
ENSDART00000126991 |
ascl1b
|
achaete-scute family bHLH transcription factor 1b |
| chr3_-_12026741 | 0.80 |
ENSDART00000132238
|
cfap70
|
cilia and flagella associated protein 70 |
| chr19_-_7115229 | 0.76 |
ENSDART00000001930
|
psmb13a
|
proteasome subunit beta 13a |
| chr11_-_18705303 | 0.75 |
ENSDART00000059732
|
id1
|
inhibitor of DNA binding 1 |
| chr7_+_65673885 | 0.74 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr25_+_245018 | 0.74 |
ENSDART00000155344
|
zgc:92481
|
zgc:92481 |
| chr11_-_38492269 | 0.69 |
ENSDART00000065613
|
elk4
|
ELK4, ETS-domain protein |
| chr11_-_38492564 | 0.69 |
ENSDART00000102850
|
elk4
|
ELK4, ETS-domain protein |
| chr1_-_44710937 | 0.67 |
ENSDART00000139036
|
si:dkey-28b4.7
|
si:dkey-28b4.7 |
| chr6_-_7718200 | 0.66 |
ENSDART00000189664
ENSDART00000114282 |
rpsa
|
ribosomal protein SA |
| chr19_+_7115223 | 0.65 |
ENSDART00000001359
|
psmb12
|
proteasome subunit beta 12 |
| chr18_-_18543358 | 0.62 |
ENSDART00000126460
|
il34
|
interleukin 34 |
| chr8_+_16004551 | 0.58 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr23_-_30787932 | 0.58 |
ENSDART00000135771
|
myt1a
|
myelin transcription factor 1a |
| chr4_+_25558849 | 0.52 |
ENSDART00000113663
ENSDART00000100755 ENSDART00000111416 ENSDART00000127840 ENSDART00000168618 ENSDART00000111820 ENSDART00000113866 ENSDART00000110107 ENSDART00000111344 ENSDART00000108548 |
zgc:195175
|
zgc:195175 |
| chr11_-_29623380 | 0.52 |
ENSDART00000162587
ENSDART00000193935 ENSDART00000191646 |
chd5
|
chromodomain helicase DNA binding protein 5 |
| chr14_-_4121052 | 0.51 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr12_+_5189776 | 0.51 |
ENSDART00000081298
|
lgi1b
|
leucine-rich, glioma inactivated 1b |
| chr3_+_24618012 | 0.50 |
ENSDART00000111997
|
zgc:171506
|
zgc:171506 |
| chr5_+_59494079 | 0.50 |
ENSDART00000148727
|
gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr11_-_34628789 | 0.48 |
ENSDART00000192433
|
mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3 |
| chr16_-_9802449 | 0.47 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr23_+_642001 | 0.46 |
ENSDART00000030643
ENSDART00000124850 |
irf10
|
interferon regulatory factor 10 |
| chr16_-_51271962 | 0.46 |
ENSDART00000164021
ENSDART00000046420 |
serpinb1l1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1, like 1 |
| chr19_+_12350069 | 0.45 |
ENSDART00000052240
|
psmg2
|
proteasome (prosome, macropain) assembly chaperone 2 |
| chr12_+_20641102 | 0.45 |
ENSDART00000152964
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr16_-_9802998 | 0.44 |
ENSDART00000154217
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr25_+_4979446 | 0.41 |
ENSDART00000154131
ENSDART00000155537 |
si:ch73-265h17.1
|
si:ch73-265h17.1 |
| chr10_+_10210455 | 0.41 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr1_-_45636362 | 0.39 |
ENSDART00000181757
|
sept3
|
septin 3 |
| chr2_+_33051730 | 0.39 |
ENSDART00000177902
ENSDART00000187903 |
rnf220a
|
ring finger protein 220a |
| chr22_+_1683185 | 0.38 |
ENSDART00000158109
|
si:dkey-1b17.6
|
si:dkey-1b17.6 |
| chr16_-_43344859 | 0.37 |
ENSDART00000058680
|
psma2
|
proteasome subunit alpha 2 |
| chr22_-_18164671 | 0.37 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr7_+_11197940 | 0.37 |
ENSDART00000081346
|
cemip
|
cell migration inducing protein, hyaluronan binding |
| chr14_+_35806605 | 0.36 |
ENSDART00000173093
|
gria2b
|
glutamate receptor, ionotropic, AMPA 2b |
| chr1_+_31942961 | 0.36 |
ENSDART00000007522
|
anos1a
|
anosmin 1a |
| chr23_+_642395 | 0.36 |
ENSDART00000186995
|
irf10
|
interferon regulatory factor 10 |
| chr2_-_19520324 | 0.35 |
ENSDART00000079877
|
pimr52
|
Pim proto-oncogene, serine/threonine kinase, related 52 |
| chr13_-_1151410 | 0.35 |
ENSDART00000007231
|
psmb1
|
proteasome subunit beta 1 |
| chr14_-_4120636 | 0.34 |
ENSDART00000059230
|
irf2
|
interferon regulatory factor 2 |
| chr3_+_59935606 | 0.34 |
ENSDART00000154157
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr16_+_43344475 | 0.33 |
ENSDART00000085282
|
mrpl32
|
mitochondrial ribosomal protein L32 |
| chr4_-_77216726 | 0.32 |
ENSDART00000099943
|
psmb10
|
proteasome subunit beta 10 |
| chr2_-_16254782 | 0.32 |
ENSDART00000133708
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr18_+_14693682 | 0.32 |
ENSDART00000132249
|
uri1
|
URI1, prefoldin-like chaperone |
| chr4_-_20208166 | 0.32 |
ENSDART00000066891
|
gsap
|
gamma-secretase activating protein |
| chr2_-_24369087 | 0.32 |
ENSDART00000081237
|
plvapa
|
plasmalemma vesicle associated protein a |
| chr20_+_47434709 | 0.32 |
ENSDART00000067776
|
rab10
|
RAB10, member RAS oncogene family |
| chr4_-_74367912 | 0.32 |
ENSDART00000174199
ENSDART00000165257 |
ptprb
|
protein tyrosine phosphatase, receptor type, b |
| chr5_+_51443009 | 0.31 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr17_+_25955003 | 0.31 |
ENSDART00000156029
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr1_-_7603734 | 0.30 |
ENSDART00000009315
|
mxb
|
myxovirus (influenza) resistance B |
| chr21_+_5636008 | 0.30 |
ENSDART00000158385
|
shroom3
|
shroom family member 3 |
| chr9_-_8454060 | 0.29 |
ENSDART00000110158
|
irs2b
|
insulin receptor substrate 2b |
| chr2_+_21486529 | 0.29 |
ENSDART00000047468
|
inhbab
|
inhibin, beta Ab |
| chr5_+_13373593 | 0.29 |
ENSDART00000051668
ENSDART00000183883 |
ccl19a.2
|
chemokine (C-C motif) ligand 19a, tandem duplicate 2 |
| chr11_-_36350421 | 0.29 |
ENSDART00000141477
|
psma5
|
proteasome subunit alpha 5 |
| chr10_+_28587446 | 0.29 |
ENSDART00000030138
ENSDART00000137508 |
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr7_-_37563883 | 0.28 |
ENSDART00000148805
|
nod2
|
nucleotide-binding oligomerization domain containing 2 |
| chr8_+_39998467 | 0.28 |
ENSDART00000073782
ENSDART00000134452 |
ggt5a
|
gamma-glutamyltransferase 5a |
| chr7_+_48870496 | 0.28 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr21_+_5169154 | 0.27 |
ENSDART00000102559
|
zgc:122979
|
zgc:122979 |
| chr5_-_43935460 | 0.25 |
ENSDART00000166152
ENSDART00000188969 |
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr1_+_31706668 | 0.24 |
ENSDART00000057879
|
as3mt
|
arsenite methyltransferase |
| chr22_-_18164835 | 0.24 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr2_+_33052169 | 0.23 |
ENSDART00000180008
|
rnf220a
|
ring finger protein 220a |
| chr22_-_23748284 | 0.23 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr21_-_7781555 | 0.22 |
ENSDART00000084380
ENSDART00000189131 |
aggf1
|
angiogenic factor with G patch and FHA domains 1 |
| chr7_+_36552725 | 0.22 |
ENSDART00000173682
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr20_-_20402500 | 0.22 |
ENSDART00000190747
ENSDART00000185065 |
prkchb
|
protein kinase C, eta, b |
| chr16_-_50181206 | 0.21 |
ENSDART00000148478
|
lim2.5
|
lens intrinsic membrane protein 2.5 |
| chr11_+_21910752 | 0.21 |
ENSDART00000114288
|
foxp4
|
forkhead box P4 |
| chr1_+_59154521 | 0.21 |
ENSDART00000130089
ENSDART00000152456 |
soul5l
|
heme-binding protein soul5, like |
| chr3_+_1942219 | 0.21 |
ENSDART00000114520
|
zgc:165583
|
zgc:165583 |
| chr7_-_8881514 | 0.21 |
ENSDART00000081620
|
vax2
|
ventral anterior homeobox 2 |
| chr16_+_5408748 | 0.21 |
ENSDART00000160008
ENSDART00000014024 |
plecb
|
plectin b |
| chr1_-_54938137 | 0.21 |
ENSDART00000147212
|
crtac1a
|
cartilage acidic protein 1a |
| chr4_+_34417403 | 0.21 |
ENSDART00000186032
ENSDART00000167909 |
si:ch211-246b8.2
|
si:ch211-246b8.2 |
| chr4_-_76370630 | 0.20 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr23_+_33718602 | 0.20 |
ENSDART00000024695
|
dazap2
|
DAZ associated protein 2 |
| chr12_+_17042754 | 0.20 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr25_+_8955530 | 0.20 |
ENSDART00000156444
|
si:ch211-256a21.4
|
si:ch211-256a21.4 |
| chr17_+_45404758 | 0.19 |
ENSDART00000147557
|
tagapa
|
T cell activation RhoGTPase activating protein a |
| chr5_-_41645058 | 0.19 |
ENSDART00000051092
|
riok2
|
RIO kinase 2 (yeast) |
| chr8_+_16004154 | 0.19 |
ENSDART00000134787
ENSDART00000172510 ENSDART00000141173 |
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr6_-_10780698 | 0.19 |
ENSDART00000151714
|
gpr155b
|
G protein-coupled receptor 155b |
| chr13_-_43637371 | 0.18 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr3_+_47322494 | 0.18 |
ENSDART00000102202
|
ppap2d
|
phosphatidic acid phosphatase type 2D |
| chr20_-_26929685 | 0.17 |
ENSDART00000132556
|
ftr79
|
finTRIM family, member 79 |
| chr23_-_37835794 | 0.17 |
ENSDART00000137358
|
cd40
|
CD40 molecule, TNF receptor superfamily member 5 |
| chr4_+_17524330 | 0.17 |
ENSDART00000172169
ENSDART00000021610 |
recql
|
RecQ helicase-like |
| chr4_+_5132951 | 0.17 |
ENSDART00000103279
|
ccnd2b
|
cyclin D2, b |
| chr16_+_13855039 | 0.16 |
ENSDART00000113764
ENSDART00000143983 |
zgc:174888
|
zgc:174888 |
| chr2_+_30969029 | 0.16 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr16_-_31686602 | 0.16 |
ENSDART00000170357
|
c1s
|
complement component 1, s subcomponent |
| chr6_-_39313027 | 0.16 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr1_+_53377432 | 0.15 |
ENSDART00000177581
|
ucp1
|
uncoupling protein 1 |
| chr5_-_42904329 | 0.15 |
ENSDART00000112807
|
cxcl20
|
chemokine (C-X-C motif) ligand 20 |
| chr1_-_21599219 | 0.14 |
ENSDART00000148327
|
adamtsl7
|
ADAMTS-like 7 |
| chr6_-_8580857 | 0.14 |
ENSDART00000138858
ENSDART00000041142 |
myh11a
|
myosin, heavy chain 11a, smooth muscle |
| chr4_-_6373735 | 0.14 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr10_+_17776981 | 0.13 |
ENSDART00000141693
|
ccl19b
|
chemokine (C-C motif) ligand 19b |
| chr23_-_3703569 | 0.13 |
ENSDART00000143731
|
pacsin1a
|
protein kinase C and casein kinase substrate in neurons 1a |
| chr15_-_41438821 | 0.13 |
ENSDART00000136952
|
nlrc8
|
NLR family CARD domain containing 8 |
| chr21_-_20711739 | 0.13 |
ENSDART00000190918
|
BX530077.1
|
|
| chr16_-_25535430 | 0.13 |
ENSDART00000077420
|
dtnbp1a
|
dystrobrevin binding protein 1a |
| chr22_-_23668356 | 0.12 |
ENSDART00000167106
ENSDART00000159622 ENSDART00000163228 |
cfh
|
complement factor H |
| chr11_-_44484952 | 0.12 |
ENSDART00000166674
ENSDART00000188016 |
mfn1b
|
mitofusin 1b |
| chr2_+_38608290 | 0.12 |
ENSDART00000159066
|
cdh24b
|
cadherin 24, type 2b |
| chr20_-_25412181 | 0.12 |
ENSDART00000062780
|
itsn2a
|
intersectin 2a |
| chr11_-_44485700 | 0.12 |
ENSDART00000186613
ENSDART00000172828 ENSDART00000113222 |
mfn1b
|
mitofusin 1b |
| chr3_-_7974028 | 0.12 |
ENSDART00000144459
|
trim35-22
|
tripartite motif containing 35-22 |
| chr11_-_21528056 | 0.12 |
ENSDART00000181626
|
srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr15_+_17752928 | 0.12 |
ENSDART00000155314
|
si:ch211-213d14.2
|
si:ch211-213d14.2 |
| chr21_+_30306369 | 0.12 |
ENSDART00000145050
ENSDART00000059420 |
lcp2a
|
lymphocyte cytosolic protein 2a |
| chr12_+_20641471 | 0.11 |
ENSDART00000133654
|
calcoco2
|
calcium binding and coiled-coil domain 2 |
| chr13_+_30912117 | 0.11 |
ENSDART00000133138
|
drgx
|
dorsal root ganglia homeobox |
| chr20_+_4793516 | 0.11 |
ENSDART00000053875
|
lgals8a
|
galectin 8a |
| chr11_-_26040594 | 0.11 |
ENSDART00000144115
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr23_-_6982966 | 0.11 |
ENSDART00000033774
ENSDART00000169965 |
edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr1_+_20069535 | 0.10 |
ENSDART00000088653
|
prss12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr13_+_30912385 | 0.10 |
ENSDART00000182642
|
drgx
|
dorsal root ganglia homeobox |
| chr5_+_32007627 | 0.10 |
ENSDART00000183061
|
scai
|
suppressor of cancer cell invasion |
| chr20_-_26936887 | 0.10 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr4_+_35130120 | 0.10 |
ENSDART00000192362
ENSDART00000181204 |
si:dkey-279j5.1
|
si:dkey-279j5.1 |
| chr7_-_3693536 | 0.10 |
ENSDART00000126381
|
si:ch211-282j17.13
|
si:ch211-282j17.13 |
| chr4_+_38550788 | 0.08 |
ENSDART00000157412
|
si:ch211-209n20.1
|
si:ch211-209n20.1 |
| chr17_-_19784035 | 0.08 |
ENSDART00000190209
|
fmn2a
|
formin 2a |
| chr8_-_17980317 | 0.08 |
ENSDART00000129148
|
tnni3k
|
TNNI3 interacting kinase |
| chr20_-_26383368 | 0.08 |
ENSDART00000024518
ENSDART00000087844 |
esr1
|
estrogen receptor 1 |
| chr12_-_16380593 | 0.08 |
ENSDART00000152133
|
hectd2
|
HECT domain containing 2 |
| chr19_+_23296616 | 0.08 |
ENSDART00000134567
|
irgf1
|
immunity-related GTPase family, f1 |
| chr4_+_43522680 | 0.08 |
ENSDART00000182252
|
si:dkeyp-53e4.4
|
si:dkeyp-53e4.4 |
| chr5_-_38197080 | 0.08 |
ENSDART00000140708
|
si:ch211-284e13.9
|
si:ch211-284e13.9 |
| chr3_-_11040575 | 0.07 |
ENSDART00000159802
|
CR382337.2
|
|
| chr5_-_43935119 | 0.07 |
ENSDART00000142271
|
si:ch211-204c21.1
|
si:ch211-204c21.1 |
| chr17_+_39242437 | 0.07 |
ENSDART00000156138
ENSDART00000128863 |
zgc:174356
|
zgc:174356 |
| chr9_+_33267211 | 0.07 |
ENSDART00000025635
|
usp9
|
ubiquitin specific peptidase 9 |
| chr4_-_71177920 | 0.07 |
ENSDART00000158287
|
si:dkey-193i10.1
|
si:dkey-193i10.1 |
| chr1_+_44710955 | 0.07 |
ENSDART00000131296
ENSDART00000142187 |
ssrp1b
|
structure specific recognition protein 1b |
| chr5_-_46980651 | 0.07 |
ENSDART00000181022
ENSDART00000168038 |
edil3a
|
EGF-like repeats and discoidin I-like domains 3a |
| chr16_-_9383629 | 0.07 |
ENSDART00000084264
ENSDART00000166958 |
adcy2a
|
adenylate cyclase 2a |
| chr5_+_21211135 | 0.06 |
ENSDART00000088492
|
bmp10
|
bone morphogenetic protein 10 |
| chr20_+_4793790 | 0.06 |
ENSDART00000153486
|
lgals8a
|
galectin 8a |
| chr24_-_20915050 | 0.06 |
ENSDART00000133763
|
si:ch211-161h7.5
|
si:ch211-161h7.5 |
| chr7_+_26167420 | 0.06 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr12_-_4028079 | 0.06 |
ENSDART00000128676
|
si:ch211-180a12.2
|
si:ch211-180a12.2 |
| chr2_-_43850637 | 0.06 |
ENSDART00000136077
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr12_-_16380242 | 0.06 |
ENSDART00000168105
|
hectd2
|
HECT domain containing 2 |
| chr9_-_15208129 | 0.06 |
ENSDART00000137043
ENSDART00000131512 |
pard3bb
|
par-3 family cell polarity regulator beta b |
| chr23_-_27123171 | 0.05 |
ENSDART00000023218
ENSDART00000146467 |
stat6
|
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr11_+_37144328 | 0.05 |
ENSDART00000162830
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr3_+_39663987 | 0.05 |
ENSDART00000184614
ENSDART00000184573 ENSDART00000183127 |
epn2
|
epsin 2 |
| chr8_-_24135171 | 0.05 |
ENSDART00000112926
|
adora1b
|
adenosine A1 receptor b |
| chr5_-_50781623 | 0.05 |
ENSDART00000114950
|
zgc:194908
|
zgc:194908 |
| chr5_+_45895791 | 0.05 |
ENSDART00000141039
|
polk
|
polymerase (DNA directed) kappa |
| chr18_-_16792561 | 0.05 |
ENSDART00000145546
|
ampd3b
|
adenosine monophosphate deaminase 3b |
| chr6_-_58149165 | 0.05 |
ENSDART00000170752
|
tox2
|
TOX high mobility group box family member 2 |
| chr4_+_1461079 | 0.05 |
ENSDART00000082383
|
trim35-14
|
tripartite motif containing 35-14 |
| chr22_+_36719821 | 0.04 |
ENSDART00000149093
|
TRIM35 (1 of many)
|
si:ch1073-324l1.1 |
| chr21_-_31143903 | 0.04 |
ENSDART00000111571
|
rap1gap2b
|
RAP1 GTPase activating protein 2b |
| chr19_+_2275019 | 0.04 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr3_-_8035737 | 0.04 |
ENSDART00000170642
|
trim35-19
|
tripartite motif containing 35-19 |
| chr1_+_57787642 | 0.04 |
ENSDART00000127091
|
si:dkey-1c7.2
|
si:dkey-1c7.2 |
| chr24_+_24285751 | 0.04 |
ENSDART00000122294
|
ythdf3
|
YTH N(6)-methyladenosine RNA binding protein 3 |
| chr4_-_22338906 | 0.03 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr8_+_39570615 | 0.03 |
ENSDART00000142557
|
lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr22_+_19230434 | 0.03 |
ENSDART00000132929
|
si:dkey-21e2.8
|
si:dkey-21e2.8 |
| chr7_+_17456686 | 0.03 |
ENSDART00000113472
|
nitr1b
|
novel immune-type receptor 1b |
| chr19_+_18739085 | 0.03 |
ENSDART00000188868
|
spaca4l
|
sperm acrosome associated 4 like |
| chr4_-_37696010 | 0.03 |
ENSDART00000169984
|
si:dkey-3p4.1
|
si:dkey-3p4.1 |
| chr13_+_21804058 | 0.03 |
ENSDART00000140307
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr19_-_10971230 | 0.03 |
ENSDART00000166196
|
LO018584.1
|
|
| chr12_-_44070043 | 0.03 |
ENSDART00000180059
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr1_-_8966046 | 0.03 |
ENSDART00000136198
|
socs1b
|
suppressor of cytokine signaling 1b |
| chr5_-_30176349 | 0.03 |
ENSDART00000186584
|
adamts8a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8a |
| chr4_-_32466676 | 0.03 |
ENSDART00000193347
ENSDART00000169396 ENSDART00000145784 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr22_+_1508545 | 0.03 |
ENSDART00000164814
|
zgc:173486
|
zgc:173486 |
| chr4_+_70791887 | 0.02 |
ENSDART00000164234
|
si:ch211-127b6.2
|
si:ch211-127b6.2 |
| chr5_-_64103863 | 0.02 |
ENSDART00000135014
ENSDART00000083684 |
pappab
|
pregnancy-associated plasma protein A, pappalysin 1b |
| chr5_-_44286987 | 0.02 |
ENSDART00000184112
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr14_+_21754521 | 0.02 |
ENSDART00000111839
|
kdm2ab
|
lysine (K)-specific demethylase 2Ab |
| chr4_-_46948863 | 0.02 |
ENSDART00000168835
|
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr22_-_10019768 | 0.02 |
ENSDART00000162077
|
BX324216.2
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm1a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:1900408 | negative regulation of cellular response to oxidative stress(GO:1900408) negative regulation of response to oxidative stress(GO:1902883) regulation of oxidative stress-induced cell death(GO:1903201) negative regulation of oxidative stress-induced cell death(GO:1903202) |
| 0.2 | 0.6 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 3.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.2 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 2.1 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 0.2 | GO:0035666 | response to protozoan(GO:0001562) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) defense response to protozoan(GO:0042832) regulation of isotype switching(GO:0045191) positive regulation of isotype switching(GO:0045830) |
| 0.1 | 0.3 | GO:0019370 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.3 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.3 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 1.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.3 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.7 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.1 | GO:0060155 | secretory granule organization(GO:0033363) platelet dense granule organization(GO:0060155) |
| 0.0 | 0.7 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 0.2 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.4 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 0.1 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 1.9 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.2 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.0 | 0.2 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.1 | GO:0002858 | response to tumor cell(GO:0002347) natural killer cell cytokine production(GO:0002370) immune response to tumor cell(GO:0002418) natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell cytokine production(GO:0002727) positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of response to biotic stimulus(GO:0002833) regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.4 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.3 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 0.1 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.3 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.7 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.9 | GO:0010950 | positive regulation of endopeptidase activity(GO:0010950) positive regulation of peptidase activity(GO:0010952) |
| 0.0 | 0.6 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.9 | GO:0021575 | hindbrain morphogenesis(GO:0021575) |
| 0.0 | 0.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.3 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.1 | GO:1900407 | regulation of cellular response to oxidative stress(GO:1900407) |
| 0.0 | 0.3 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.3 | GO:0001843 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) |
| 0.0 | 1.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.5 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.5 | GO:0048854 | brain morphogenesis(GO:0048854) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 3.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 2.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.4 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.8 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 0.9 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 2.1 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.3 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.2 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0005153 | interleukin-8 receptor binding(GO:0005153) |
| 0.0 | 0.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 1.9 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.9 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 1.2 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |