Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
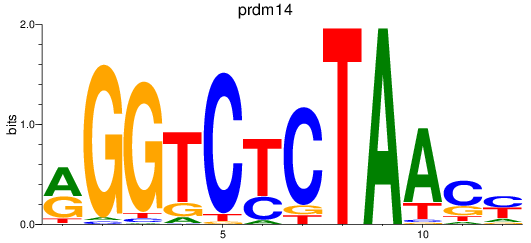
Results for prdm14
Z-value: 0.36
Transcription factors associated with prdm14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
prdm14
|
ENSDARG00000045371 | PR domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| prdm14 | dr11_v1_chr24_+_14451404_14451404 | -0.75 | 3.4e-04 | Click! |
Activity profile of prdm14 motif
Sorted Z-values of prdm14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_34794829 | 0.98 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr20_+_51061695 | 0.98 |
ENSDART00000134416
|
im:7140055
|
im:7140055 |
| chr5_+_37903790 | 0.93 |
ENSDART00000162470
|
tmprss4b
|
transmembrane protease, serine 4b |
| chr23_+_9522942 | 0.87 |
ENSDART00000137751
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr23_+_21261313 | 0.85 |
ENSDART00000104268
ENSDART00000159046 |
emc1
|
ER membrane protein complex subunit 1 |
| chr14_+_35428152 | 0.80 |
ENSDART00000172597
|
sytl4
|
synaptotagmin-like 4 |
| chr23_+_9522781 | 0.78 |
ENSDART00000136486
|
osbpl2b
|
oxysterol binding protein-like 2b |
| chr10_-_32877348 | 0.77 |
ENSDART00000018977
ENSDART00000133421 |
rabgef1
|
RAB guanine nucleotide exchange factor (GEF) 1 |
| chr1_+_46509176 | 0.68 |
ENSDART00000166028
|
mcf2la
|
mcf.2 cell line derived transforming sequence-like a |
| chr17_+_25871304 | 0.66 |
ENSDART00000185143
|
wapla
|
WAPL cohesin release factor a |
| chr17_+_12075805 | 0.60 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr5_+_28271412 | 0.58 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr8_-_25716074 | 0.55 |
ENSDART00000007482
|
tspy
|
testis specific protein, Y-linked |
| chr9_-_34945566 | 0.52 |
ENSDART00000131908
ENSDART00000059861 |
dcun1d2a
|
DCN1, defective in cullin neddylation 1, domain containing 2a |
| chr13_+_49727333 | 0.52 |
ENSDART00000168799
ENSDART00000037559 |
ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr11_+_31380495 | 0.48 |
ENSDART00000185073
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr21_-_14692119 | 0.46 |
ENSDART00000123047
|
ehmt1b
|
euchromatic histone-lysine N-methyltransferase 1b |
| chr20_-_4049862 | 0.44 |
ENSDART00000158057
|
sprtn
|
SprT-like N-terminal domain |
| chr8_+_47677208 | 0.42 |
ENSDART00000123254
|
dpp9
|
dipeptidyl-peptidase 9 |
| chr21_-_27272657 | 0.42 |
ENSDART00000040754
ENSDART00000175009 |
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr5_+_51833305 | 0.42 |
ENSDART00000165276
ENSDART00000166443 |
papd4
|
PAP associated domain containing 4 |
| chr8_-_25566347 | 0.39 |
ENSDART00000138289
ENSDART00000078022 |
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr1_+_27690 | 0.38 |
ENSDART00000162928
|
eed
|
embryonic ectoderm development |
| chr17_+_30843881 | 0.35 |
ENSDART00000149600
ENSDART00000148547 |
tpp1
|
tripeptidyl peptidase I |
| chr2_-_32826108 | 0.35 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr5_-_69482891 | 0.34 |
ENSDART00000109487
|
CABZ01032476.1
|
|
| chr2_-_37458527 | 0.34 |
ENSDART00000146820
|
si:dkey-57k2.7
|
si:dkey-57k2.7 |
| chr5_+_51833132 | 0.32 |
ENSDART00000167491
|
papd4
|
PAP associated domain containing 4 |
| chr17_+_27162367 | 0.32 |
ENSDART00000193345
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr9_+_8364553 | 0.31 |
ENSDART00000190713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr5_-_67115872 | 0.30 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr21_-_27273147 | 0.30 |
ENSDART00000143239
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr23_-_27442544 | 0.30 |
ENSDART00000019521
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr22_-_20342260 | 0.26 |
ENSDART00000161610
ENSDART00000165667 |
tcf3b
|
transcription factor 3b |
| chr11_+_13642157 | 0.24 |
ENSDART00000060251
|
wdr18
|
WD repeat domain 18 |
| chr7_+_13491452 | 0.22 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr16_+_32014552 | 0.19 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr10_+_573667 | 0.19 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr6_-_39919982 | 0.19 |
ENSDART00000065091
ENSDART00000064903 |
sumf1
|
sulfatase modifying factor 1 |
| chr12_-_4435303 | 0.19 |
ENSDART00000159007
|
si:ch211-173d10.1
|
si:ch211-173d10.1 |
| chr10_-_33343244 | 0.19 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr23_+_5104743 | 0.19 |
ENSDART00000123191
|
ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr7_+_51324834 | 0.16 |
ENSDART00000114429
|
usp12b
|
ubiquitin specific peptidase 12b |
| chr4_-_17353100 | 0.15 |
ENSDART00000134467
ENSDART00000189019 |
parpbp
|
PARP1 binding protein |
| chr15_-_34930727 | 0.14 |
ENSDART00000179723
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr11_-_23080970 | 0.14 |
ENSDART00000127791
|
atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr3_-_25268751 | 0.13 |
ENSDART00000139423
|
mgat3a
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase a |
| chr13_+_18321140 | 0.13 |
ENSDART00000180947
|
eif4e1c
|
eukaryotic translation initiation factor 4E family member 1c |
| chr18_-_25646286 | 0.12 |
ENSDART00000099511
ENSDART00000186890 |
si:ch211-13k12.2
|
si:ch211-13k12.2 |
| chr10_+_29849497 | 0.12 |
ENSDART00000099994
ENSDART00000132212 |
hspa8
|
heat shock protein 8 |
| chr8_-_49285801 | 0.11 |
ENSDART00000113159
|
prickle3
|
prickle homolog 3 |
| chr6_-_24384654 | 0.11 |
ENSDART00000164723
|
brdt
|
bromodomain, testis-specific |
| chr2_-_36819624 | 0.10 |
ENSDART00000140844
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr11_-_23332592 | 0.09 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr24_+_26402110 | 0.07 |
ENSDART00000133684
|
si:ch211-230g15.5
|
si:ch211-230g15.5 |
| chr24_-_30275735 | 0.07 |
ENSDART00000168723
|
snx7
|
sorting nexin 7 |
| chr17_+_28675120 | 0.04 |
ENSDART00000159067
|
hectd1
|
HECT domain containing 1 |
| chr6_-_24143923 | 0.04 |
ENSDART00000157948
|
si:ch73-389b16.1
|
si:ch73-389b16.1 |
| chr22_+_16308806 | 0.04 |
ENSDART00000162685
|
lrrc39
|
leucine rich repeat containing 39 |
| chr6_-_21534301 | 0.03 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr12_+_39685485 | 0.03 |
ENSDART00000163403
|
LO017650.1
|
|
| chr9_-_40073255 | 0.02 |
ENSDART00000189139
|
IKZF2
|
si:zfos-1425h8.1 |
| chr20_-_46467280 | 0.02 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr15_-_37674120 | 0.02 |
ENSDART00000059568
|
si:ch211-137j23.6
|
si:ch211-137j23.6 |
| chr21_-_43949208 | 0.01 |
ENSDART00000150983
|
camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr20_+_22220988 | 0.01 |
ENSDART00000049204
|
kdr
|
kinase insert domain receptor (a type III receptor tyrosine kinase) |
| chr22_-_9890386 | 0.01 |
ENSDART00000132304
|
si:dkey-253d23.5
|
si:dkey-253d23.5 |
| chr13_+_4871886 | 0.01 |
ENSDART00000132301
|
micu1
|
mitochondrial calcium uptake 1 |
| chr4_+_77681389 | 0.01 |
ENSDART00000099727
|
gbp4
|
guanylate binding protein 4 |
| chr22_+_2680895 | 0.01 |
ENSDART00000082245
|
zgc:194221
|
zgc:194221 |
| chr7_-_17780048 | 0.01 |
ENSDART00000183336
|
si:dkey-106g10.7
|
si:dkey-106g10.7 |
| chr11_+_13159988 | 0.00 |
ENSDART00000064176
|
mob3c
|
MOB kinase activator 3C |
| chr18_+_44768829 | 0.00 |
ENSDART00000016271
|
ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr12_-_36268723 | 0.00 |
ENSDART00000113740
|
kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
Network of associatons between targets according to the STRING database.
First level regulatory network of prdm14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.2 | 0.6 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.1 | 0.4 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.1 | GO:0051037 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.5 | GO:0045116 | protein neddylation(GO:0045116) positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.3 | GO:2000758 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0048260 | positive regulation of receptor-mediated endocytosis(GO:0048260) protein localization to early endosome(GO:1902946) |
| 0.0 | 0.2 | GO:0090329 | regulation of DNA-dependent DNA replication(GO:0090329) |
| 0.0 | 0.4 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.5 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 0.2 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.8 | GO:0016571 | histone methylation(GO:0016571) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.5 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.6 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |