Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
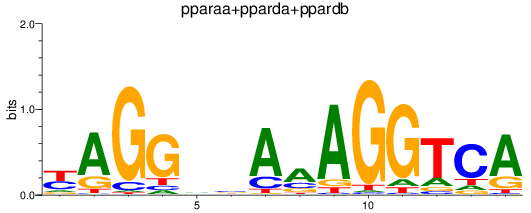
Results for pparaa+pparda+ppardb
Z-value: 0.51
Transcription factors associated with pparaa+pparda+ppardb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ppardb
|
ENSDARG00000009473 | peroxisome proliferator-activated receptor delta b |
|
pparaa
|
ENSDARG00000031777 | peroxisome proliferator-activated receptor alpha a |
|
pparda
|
ENSDARG00000044525 | peroxisome proliferator-activated receptor delta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pparaa | dr11_v1_chr4_-_18635005_18635005 | 0.91 | 1.2e-07 | Click! |
| ppardb | dr11_v1_chr8_+_23711842_23711842 | 0.75 | 3.2e-04 | Click! |
| pparda | dr11_v1_chr22_+_1006573_1006573 | 0.56 | 1.5e-02 | Click! |
Activity profile of pparaa+pparda+ppardb motif
Sorted Z-values of pparaa+pparda+ppardb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_12408109 | 1.63 |
ENSDART00000155509
|
ankef1b
|
ankyrin repeat and EF-hand domain containing 1b |
| chr25_+_21829777 | 1.32 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr10_-_44027391 | 1.31 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr2_-_42960353 | 1.07 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr19_+_40861853 | 1.05 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr5_-_323712 | 1.04 |
ENSDART00000188793
|
HOOK3
|
hook microtubule tethering protein 3 |
| chr25_+_5288665 | 1.00 |
ENSDART00000169540
|
CABZ01039861.1
|
|
| chr23_-_39636195 | 0.92 |
ENSDART00000144439
|
vwa1
|
von Willebrand factor A domain containing 1 |
| chr5_-_36549024 | 0.83 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr3_-_53486169 | 0.83 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr13_-_37127970 | 0.81 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr8_+_999421 | 0.81 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr7_-_24046999 | 0.77 |
ENSDART00000144616
ENSDART00000124653 ENSDART00000127813 |
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr22_+_22888 | 0.75 |
ENSDART00000082471
|
mfap2
|
microfibril associated protein 2 |
| chr20_+_54738210 | 0.73 |
ENSDART00000151399
|
pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr5_-_30984010 | 0.72 |
ENSDART00000182367
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr18_+_16744307 | 0.72 |
ENSDART00000179872
ENSDART00000133490 |
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr5_-_30984271 | 0.68 |
ENSDART00000051392
|
spns3
|
spinster homolog 3 (Drosophila) |
| chr13_+_28819768 | 0.65 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr24_+_9590188 | 0.62 |
ENSDART00000137092
|
si:dkey-96n2.1
|
si:dkey-96n2.1 |
| chr17_+_12408188 | 0.62 |
ENSDART00000105218
|
khk
|
ketohexokinase |
| chr17_+_32374876 | 0.59 |
ENSDART00000183851
|
ywhaqb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide b |
| chr21_+_26720803 | 0.59 |
ENSDART00000053797
|
slc3a2b
|
solute carrier family 3 (amino acid transporter heavy chain), member 2b |
| chr12_+_7491690 | 0.58 |
ENSDART00000152564
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr12_-_684200 | 0.57 |
ENSDART00000152122
|
si:ch211-176g6.2
|
si:ch211-176g6.2 |
| chr19_+_19652439 | 0.56 |
ENSDART00000165934
|
hibadha
|
3-hydroxyisobutyrate dehydrogenase a |
| chr22_-_38274188 | 0.49 |
ENSDART00000139420
ENSDART00000015117 |
elavl2
|
ELAV like neuron-specific RNA binding protein 2 |
| chr11_+_11201096 | 0.47 |
ENSDART00000171916
ENSDART00000171521 ENSDART00000087105 ENSDART00000159603 |
myom2a
|
myomesin 2a |
| chr17_+_30545895 | 0.47 |
ENSDART00000076739
|
nhsl1a
|
NHS-like 1a |
| chr23_+_44634187 | 0.43 |
ENSDART00000143688
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr6_-_39764995 | 0.42 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr12_-_5120339 | 0.41 |
ENSDART00000168759
|
rbp4
|
retinol binding protein 4, plasma |
| chr7_-_29231686 | 0.40 |
ENSDART00000173780
|
herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr11_-_6974022 | 0.39 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr3_+_58167288 | 0.39 |
ENSDART00000155874
ENSDART00000010395 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein 2a |
| chr23_-_45504991 | 0.38 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr20_-_38836161 | 0.38 |
ENSDART00000061358
|
si:dkey-221h15.4
|
si:dkey-221h15.4 |
| chr6_+_60055168 | 0.38 |
ENSDART00000008752
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr23_+_44633858 | 0.32 |
ENSDART00000180728
|
si:ch73-265d7.2
|
si:ch73-265d7.2 |
| chr14_-_48103207 | 0.31 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr5_+_38276582 | 0.30 |
ENSDART00000158532
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr20_-_26937453 | 0.29 |
ENSDART00000139756
|
ftr97
|
finTRIM family, member 97 |
| chr6_-_39765546 | 0.28 |
ENSDART00000185767
|
pfkmb
|
phosphofructokinase, muscle b |
| chr6_-_18393476 | 0.28 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr8_-_979735 | 0.28 |
ENSDART00000149612
|
znf366
|
zinc finger protein 366 |
| chr12_-_5120175 | 0.26 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr12_-_31457301 | 0.26 |
ENSDART00000043887
ENSDART00000148603 |
acsl5
|
acyl-CoA synthetase long chain family member 5 |
| chr20_-_24443680 | 0.24 |
ENSDART00000191337
|
CR385053.3
|
|
| chr5_+_43870389 | 0.24 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr24_-_1021318 | 0.23 |
ENSDART00000181403
|
ralaa
|
v-ral simian leukemia viral oncogene homolog Aa (ras related) |
| chr25_+_17405458 | 0.23 |
ENSDART00000186711
|
e2f4
|
E2F transcription factor 4 |
| chr18_+_13315739 | 0.23 |
ENSDART00000143404
|
si:ch211-260p9.3
|
si:ch211-260p9.3 |
| chr1_+_58353661 | 0.22 |
ENSDART00000140074
|
si:dkey-222h21.2
|
si:dkey-222h21.2 |
| chr10_+_20128267 | 0.22 |
ENSDART00000064615
|
dmtn
|
dematin actin binding protein |
| chr13_+_829585 | 0.22 |
ENSDART00000029051
|
gsta.2
|
glutathione S-transferase, alpha tandem duplicate 2 |
| chr17_+_12408405 | 0.22 |
ENSDART00000154827
ENSDART00000048440 ENSDART00000156429 |
khk
|
ketohexokinase |
| chr13_+_28821841 | 0.22 |
ENSDART00000179900
|
CU639469.1
|
|
| chr19_-_7043355 | 0.21 |
ENSDART00000104845
|
tapbp.1
|
TAP binding protein (tapasin), tandem duplicate 1 |
| chr19_+_3056450 | 0.19 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr12_-_6033824 | 0.19 |
ENSDART00000131301
ENSDART00000139419 ENSDART00000032050 |
g6pca.1
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 1 |
| chr19_+_7043634 | 0.19 |
ENSDART00000133954
|
mhc1uka
|
major histocompatibility complex class I UKA |
| chr25_+_17405201 | 0.19 |
ENSDART00000164349
|
e2f4
|
E2F transcription factor 4 |
| chr5_-_66301142 | 0.19 |
ENSDART00000067541
|
prlrb
|
prolactin receptor b |
| chr7_+_17356482 | 0.18 |
ENSDART00000063629
|
nitr3r.1l
|
novel immune-type receptor 3, related 1-like |
| chr5_+_58421536 | 0.17 |
ENSDART00000173410
|
ccdc15
|
coiled-coil domain containing 15 |
| chr8_-_20291922 | 0.17 |
ENSDART00000148304
|
myo1f
|
myosin IF |
| chr15_+_17030473 | 0.16 |
ENSDART00000129407
|
plin2
|
perilipin 2 |
| chr6_+_43353271 | 0.16 |
ENSDART00000191775
|
BX927362.2
|
|
| chr18_-_3456117 | 0.16 |
ENSDART00000158508
|
myo7aa
|
myosin VIIAa |
| chr20_+_43379029 | 0.16 |
ENSDART00000142486
ENSDART00000186486 |
unc93a
|
unc-93 homolog A |
| chr14_-_29826659 | 0.15 |
ENSDART00000138413
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr17_-_12196865 | 0.15 |
ENSDART00000154694
|
kif28
|
kinesin family member 28 |
| chr20_-_26936887 | 0.15 |
ENSDART00000160827
|
ftr79
|
finTRIM family, member 79 |
| chr4_+_67999732 | 0.14 |
ENSDART00000190597
|
si:ch211-133h13.1
|
si:ch211-133h13.1 |
| chr5_-_41860556 | 0.14 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr17_+_450956 | 0.14 |
ENSDART00000183022
ENSDART00000171386 |
zgc:194887
|
zgc:194887 |
| chr5_-_32890807 | 0.14 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr8_-_8698607 | 0.13 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr20_+_15015557 | 0.13 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr4_-_16451375 | 0.13 |
ENSDART00000192700
ENSDART00000128835 |
wu:fc23c09
|
wu:fc23c09 |
| chr20_-_52967878 | 0.12 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr13_-_31878043 | 0.12 |
ENSDART00000187720
|
syt14a
|
synaptotagmin XIVa |
| chr13_-_31878263 | 0.12 |
ENSDART00000180411
|
syt14a
|
synaptotagmin XIVa |
| chr9_+_41224100 | 0.12 |
ENSDART00000141792
|
stat1b
|
signal transducer and activator of transcription 1b |
| chr25_+_36347126 | 0.12 |
ENSDART00000152449
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr23_+_36340520 | 0.12 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr7_+_34297271 | 0.11 |
ENSDART00000180342
|
bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr17_+_30546579 | 0.11 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr24_+_2961098 | 0.11 |
ENSDART00000163760
ENSDART00000170835 |
eci2
|
enoyl-CoA delta isomerase 2 |
| chr6_-_59563597 | 0.10 |
ENSDART00000166311
|
INHBE
|
inhibin beta E subunit |
| chr7_+_48675347 | 0.10 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr1_-_58868306 | 0.10 |
ENSDART00000166615
|
dnm2b
|
dynamin 2b |
| chr19_+_32158010 | 0.10 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr16_+_28601974 | 0.10 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
| chr25_-_19420949 | 0.09 |
ENSDART00000181338
|
map1ab
|
microtubule-associated protein 1Ab |
| chr15_+_404891 | 0.09 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr3_+_30257582 | 0.09 |
ENSDART00000159497
ENSDART00000103457 ENSDART00000121883 |
mybpc2a
|
myosin binding protein C, fast type a |
| chr4_-_20243021 | 0.09 |
ENSDART00000048023
|
cacna2d4a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4a |
| chr7_-_6279138 | 0.08 |
ENSDART00000173299
|
si:ch211-220f21.2
|
si:ch211-220f21.2 |
| chr23_-_24488696 | 0.08 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr17_-_20711735 | 0.07 |
ENSDART00000150056
|
ank3b
|
ankyrin 3b |
| chr18_+_27926839 | 0.06 |
ENSDART00000191835
|
hipk3b
|
homeodomain interacting protein kinase 3b |
| chr19_+_20274944 | 0.06 |
ENSDART00000151237
|
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr20_-_34164278 | 0.06 |
ENSDART00000153128
|
hmcn1
|
hemicentin 1 |
| chr15_+_28268135 | 0.06 |
ENSDART00000152536
ENSDART00000188550 |
myo1cb
|
myosin Ic, paralog b |
| chr20_-_25877793 | 0.05 |
ENSDART00000177333
ENSDART00000184619 |
exoc1
|
exocyst complex component 1 |
| chr9_-_1939232 | 0.05 |
ENSDART00000146131
|
hoxd3a
|
homeobox D3a |
| chr7_+_11459235 | 0.04 |
ENSDART00000159611
|
il16
|
interleukin 16 |
| chr21_-_19006631 | 0.04 |
ENSDART00000080269
ENSDART00000191682 |
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr5_+_38200807 | 0.04 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr15_+_17031111 | 0.03 |
ENSDART00000175378
|
plin2
|
perilipin 2 |
| chr10_-_17284055 | 0.03 |
ENSDART00000167464
|
GNAZ
|
G protein subunit alpha z |
| chr22_+_9239831 | 0.03 |
ENSDART00000133720
|
si:ch211-250k18.5
|
si:ch211-250k18.5 |
| chr20_+_46286082 | 0.03 |
ENSDART00000073519
|
taar14f
|
trace amine associated receptor 14f |
| chr5_-_23362602 | 0.03 |
ENSDART00000137120
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr1_+_40802454 | 0.03 |
ENSDART00000193568
|
cpz
|
carboxypeptidase Z |
| chr7_+_23940933 | 0.03 |
ENSDART00000173628
|
si:dkey-183c6.7
|
si:dkey-183c6.7 |
| chr22_+_38229321 | 0.02 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr1_-_52292235 | 0.02 |
ENSDART00000132638
|
si:dkey-121b10.7
|
si:dkey-121b10.7 |
| chr6_+_60036767 | 0.02 |
ENSDART00000155009
|
tgm8
|
transglutaminase 8 |
| chr23_-_16734009 | 0.02 |
ENSDART00000125449
|
si:ch211-224l10.4
|
si:ch211-224l10.4 |
| chr19_+_2275019 | 0.02 |
ENSDART00000136138
|
itgb8
|
integrin, beta 8 |
| chr13_-_28688104 | 0.02 |
ENSDART00000133827
|
pcgf6
|
polycomb group ring finger 6 |
| chr22_-_3595439 | 0.01 |
ENSDART00000083308
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr1_-_17797802 | 0.01 |
ENSDART00000041215
|
sorbs2a
|
sorbin and SH3 domain containing 2a |
| chr5_+_36974931 | 0.01 |
ENSDART00000193063
|
gjd1a
|
gap junction protein delta 1a |
| chr16_-_29480335 | 0.01 |
ENSDART00000148930
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr18_+_15182965 | 0.00 |
ENSDART00000137018
|
si:ch73-62b13.1
|
si:ch73-62b13.1 |
| chr11_+_30981777 | 0.00 |
ENSDART00000148949
|
cacna1ab
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit, b |
| chr1_+_25848231 | 0.00 |
ENSDART00000027973
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr19_-_43552252 | 0.00 |
ENSDART00000138308
|
gpr186
|
G protein-coupled receptor 186 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pparaa+pparda+ppardb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0009750 | response to fructose(GO:0009750) |
| 0.1 | 1.3 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 0.7 | GO:0034633 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.1 | 0.4 | GO:0071548 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) short-chain fatty acid catabolic process(GO:0019626) response to dexamethasone(GO:0071548) |
| 0.1 | 0.6 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.3 | GO:0039531 | RIG-I signaling pathway(GO:0039529) regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039531) regulation of RIG-I signaling pathway(GO:0039535) |
| 0.1 | 0.3 | GO:1902001 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 1.1 | GO:0032309 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.1 | 0.7 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.6 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 1.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0008442 | 3-hydroxyisobutyrate dehydrogenase activity(GO:0008442) |
| 0.1 | 0.3 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 1.3 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.7 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.1 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.7 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 1.1 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.8 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.3 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.6 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 2.1 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.3 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.4 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |