Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
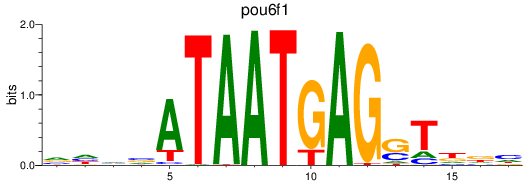
Results for pou6f1
Z-value: 0.45
Transcription factors associated with pou6f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou6f1
|
ENSDARG00000011570 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou6f1 | dr11_v1_chr23_-_33702900_33702900 | 0.88 | 1.2e-06 | Click! |
Activity profile of pou6f1 motif
Sorted Z-values of pou6f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25777425 | 1.00 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr1_-_53468160 | 0.96 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr7_-_19364813 | 0.89 |
ENSDART00000173977
|
ntn4
|
netrin 4 |
| chr16_-_29387215 | 0.74 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr21_-_13690712 | 0.65 |
ENSDART00000065817
|
pou5f3
|
POU domain, class 5, transcription factor 3 |
| chr7_+_38717624 | 0.51 |
ENSDART00000132522
|
syt13
|
synaptotagmin XIII |
| chr9_+_34151367 | 0.47 |
ENSDART00000143991
|
gpr161
|
G protein-coupled receptor 161 |
| chr15_-_931815 | 0.45 |
ENSDART00000106627
ENSDART00000102239 ENSDART00000184754 ENSDART00000186733 |
zgc:162936
|
zgc:162936 |
| chr15_-_33172246 | 0.44 |
ENSDART00000158666
|
nbeab
|
neurobeachin b |
| chr18_-_43866001 | 0.40 |
ENSDART00000150218
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr3_+_1219344 | 0.38 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr18_-_43866526 | 0.37 |
ENSDART00000111309
|
treh
|
trehalase (brush-border membrane glycoprotein) |
| chr10_-_45058886 | 0.36 |
ENSDART00000159347
|
mrps24
|
mitochondrial ribosomal protein S24 |
| chr13_+_43400443 | 0.35 |
ENSDART00000084321
|
dact2
|
dishevelled-binding antagonist of beta-catenin 2 |
| chr1_-_51710225 | 0.35 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr4_-_72609118 | 0.34 |
ENSDART00000174077
|
si:cabz01054396.2
|
si:cabz01054396.2 |
| chr2_-_21625493 | 0.30 |
ENSDART00000137169
|
zgc:55781
|
zgc:55781 |
| chr2_-_20981907 | 0.29 |
ENSDART00000113384
|
lyrm4
|
LYR motif containing 4 |
| chr18_+_34181655 | 0.29 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr18_-_20608025 | 0.27 |
ENSDART00000090156
ENSDART00000151980 |
bcl2l13
|
BCL2 like 13 |
| chr2_-_37312927 | 0.27 |
ENSDART00000141214
|
skila
|
SKI-like proto-oncogene a |
| chr5_-_39736383 | 0.27 |
ENSDART00000127123
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr24_-_25096199 | 0.26 |
ENSDART00000185076
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr2_+_29249204 | 0.26 |
ENSDART00000168957
|
cdh18a
|
cadherin 18, type 2a |
| chr14_-_28565675 | 0.26 |
ENSDART00000193961
|
insb
|
preproinsulin b |
| chr14_-_28566238 | 0.25 |
ENSDART00000172547
|
insb
|
preproinsulin b |
| chr18_+_3243292 | 0.25 |
ENSDART00000166580
|
pak1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr9_-_16218097 | 0.25 |
ENSDART00000190503
|
myo1b
|
myosin IB |
| chr7_+_13988075 | 0.24 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr15_-_5720583 | 0.24 |
ENSDART00000158034
ENSDART00000190332 ENSDART00000109599 |
urb1
|
URB1 ribosome biogenesis 1 homolog (S. cerevisiae) |
| chr16_-_25368048 | 0.24 |
ENSDART00000132445
|
rbfa
|
ribosome binding factor A |
| chr4_-_2380173 | 0.24 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr7_+_48297842 | 0.24 |
ENSDART00000052123
|
slc25a44b
|
solute carrier family 25, member 44 b |
| chr24_-_16979728 | 0.24 |
ENSDART00000005331
|
klhl15
|
kelch-like family member 15 |
| chr2_+_38892631 | 0.23 |
ENSDART00000134040
|
si:ch211-119o8.4
|
si:ch211-119o8.4 |
| chr22_-_11829436 | 0.22 |
ENSDART00000126784
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr12_+_32729470 | 0.22 |
ENSDART00000175712
|
rbfox3a
|
RNA binding fox-1 homolog 3a |
| chr2_+_29249561 | 0.20 |
ENSDART00000099157
|
cdh18a
|
cadherin 18, type 2a |
| chr1_+_50968908 | 0.19 |
ENSDART00000150353
ENSDART00000012842 |
mdh1aa
|
malate dehydrogenase 1Aa, NAD (soluble) |
| chr1_+_19649545 | 0.19 |
ENSDART00000054575
|
tmem192
|
transmembrane protein 192 |
| chr6_-_9676108 | 0.18 |
ENSDART00000169915
|
nop58
|
NOP58 ribonucleoprotein homolog (yeast) |
| chr17_-_24587686 | 0.17 |
ENSDART00000143084
|
aftphb
|
aftiphilin b |
| chr21_+_40292801 | 0.17 |
ENSDART00000174220
|
si:ch211-218m3.16
|
si:ch211-218m3.16 |
| chr25_-_8625601 | 0.17 |
ENSDART00000155280
|
GDPGP1
|
zgc:153343 |
| chr25_+_28555584 | 0.17 |
ENSDART00000157046
|
SLC15A5
|
si:ch211-190o6.3 |
| chr7_+_53498152 | 0.16 |
ENSDART00000184497
|
znf609b
|
zinc finger protein 609b |
| chr25_-_20666754 | 0.16 |
ENSDART00000158418
|
csk
|
C-terminal Src kinase |
| chr14_+_3287740 | 0.15 |
ENSDART00000186290
|
cdx1a
|
caudal type homeobox 1a |
| chr11_+_11303458 | 0.15 |
ENSDART00000162486
ENSDART00000160703 |
si:dkey-23f9.4
|
si:dkey-23f9.4 |
| chr22_-_11520405 | 0.15 |
ENSDART00000063157
|
slc26a11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr9_-_6624663 | 0.15 |
ENSDART00000092537
|
GPR45
|
si:dkeyp-118h3.5 |
| chr2_-_27619954 | 0.15 |
ENSDART00000144826
|
tgs1
|
trimethylguanosine synthase 1 |
| chr3_-_34136368 | 0.14 |
ENSDART00000136900
ENSDART00000186125 |
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr21_-_40938382 | 0.14 |
ENSDART00000008593
|
yipf5
|
Yip1 domain family, member 5 |
| chr11_-_14813029 | 0.13 |
ENSDART00000004920
ENSDART00000122645 |
sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr1_-_34335752 | 0.12 |
ENSDART00000140157
|
si:dkey-24h22.5
|
si:dkey-24h22.5 |
| chr3_-_34136778 | 0.12 |
ENSDART00000131951
|
clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr8_+_37168570 | 0.12 |
ENSDART00000098636
|
csf1b
|
colony stimulating factor 1b (macrophage) |
| chr11_+_41981959 | 0.12 |
ENSDART00000055707
|
her15.1
|
hairy and enhancer of split-related 15, tandem duplicate 1 |
| chr7_+_41147732 | 0.12 |
ENSDART00000185388
|
puf60b
|
poly-U binding splicing factor b |
| chr25_-_20666328 | 0.11 |
ENSDART00000098076
|
csk
|
C-terminal Src kinase |
| chr18_-_40773413 | 0.11 |
ENSDART00000133797
|
vaspb
|
vasodilator stimulated phosphoprotein b |
| chr14_+_46003973 | 0.11 |
ENSDART00000145241
ENSDART00000141545 |
slu7
|
SLU7 homolog, splicing factor |
| chr24_-_32408404 | 0.11 |
ENSDART00000144157
|
si:ch211-56a11.2
|
si:ch211-56a11.2 |
| chr22_+_34710473 | 0.10 |
ENSDART00000155906
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr14_-_31465905 | 0.10 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr22_+_34710036 | 0.10 |
ENSDART00000025820
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr2_-_12243213 | 0.09 |
ENSDART00000113081
|
gpr158b
|
G protein-coupled receptor 158b |
| chr21_-_30166097 | 0.09 |
ENSDART00000130676
|
hbegfb
|
heparin-binding EGF-like growth factor b |
| chr9_-_38036984 | 0.09 |
ENSDART00000134574
|
hacd2
|
3-hydroxyacyl-CoA dehydratase 2 |
| chr11_+_33628104 | 0.09 |
ENSDART00000165318
|
thsd7bb
|
thrombospondin, type I, domain containing 7Bb |
| chr17_+_8799451 | 0.09 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr18_+_33276609 | 0.09 |
ENSDART00000137208
|
v2rx3
|
vomeronasal 2 receptor, x3 |
| chr13_+_3938526 | 0.08 |
ENSDART00000012759
|
yipf3
|
Yip1 domain family, member 3 |
| chr22_-_15385442 | 0.08 |
ENSDART00000090975
|
tmem264
|
transmembrane protein 264 |
| chr13_-_46200240 | 0.08 |
ENSDART00000056984
|
ftr69
|
finTRIM family, member 69 |
| chr23_+_45860676 | 0.07 |
ENSDART00000012234
|
syap1
|
synapse associated protein 1 |
| chr4_-_75505048 | 0.07 |
ENSDART00000163665
|
si:dkey-71l4.5
|
si:dkey-71l4.5 |
| chr9_-_44953349 | 0.07 |
ENSDART00000135156
|
vil1
|
villin 1 |
| chr9_-_1469083 | 0.07 |
ENSDART00000144096
|
rbm45
|
RNA binding motif protein 45 |
| chr9_+_11034314 | 0.07 |
ENSDART00000032695
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr1_+_11992 | 0.06 |
ENSDART00000161842
|
cep97
|
centrosomal protein 97 |
| chr22_+_2793382 | 0.06 |
ENSDART00000144980
|
si:dkey-20i20.8
|
si:dkey-20i20.8 |
| chr1_+_52633367 | 0.06 |
ENSDART00000134658
|
slc44a1a
|
solute carrier family 44 (choline transporter), member 1a |
| chr1_+_33634186 | 0.06 |
ENSDART00000075626
|
htr1fb
|
5-hydroxytryptamine (serotonin) receptor 1Fb |
| chr4_-_19028861 | 0.06 |
ENSDART00000166374
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr4_+_25431990 | 0.06 |
ENSDART00000176982
|
prkcq
|
protein kinase C, theta |
| chr10_+_36345176 | 0.06 |
ENSDART00000099397
|
or105-1
|
odorant receptor, family C, subfamily 105, member 1 |
| chr21_-_7953792 | 0.06 |
ENSDART00000124486
|
si:dkey-163m14.10
|
si:dkey-163m14.10 |
| chr3_+_40796110 | 0.05 |
ENSDART00000014729
|
arpc1a
|
actin related protein 2/3 complex, subunit 1A |
| chr6_+_52938534 | 0.05 |
ENSDART00000174150
|
BX649282.2
|
|
| chr2_-_21349425 | 0.05 |
ENSDART00000171699
|
hhatla
|
hedgehog acyltransferase like, a |
| chr1_+_58535306 | 0.05 |
ENSDART00000165673
|
zgc:172091
|
zgc:172091 |
| chr4_+_14826299 | 0.05 |
ENSDART00000067035
|
shisal1a
|
shisa like 1a |
| chr4_-_31784872 | 0.05 |
ENSDART00000182911
|
si:dkey-19c16.12
|
si:dkey-19c16.12 |
| chr13_-_24825691 | 0.05 |
ENSDART00000142745
|
slka
|
STE20-like kinase a |
| chr17_-_30366705 | 0.04 |
ENSDART00000154906
|
si:ch211-139l2.1
|
si:ch211-139l2.1 |
| chr24_+_22485710 | 0.04 |
ENSDART00000146058
|
si:dkey-40h20.1
|
si:dkey-40h20.1 |
| chr2_+_29842878 | 0.04 |
ENSDART00000131376
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr2_-_57941037 | 0.04 |
ENSDART00000131420
|
si:dkeyp-68b7.5
|
si:dkeyp-68b7.5 |
| chr13_+_18533005 | 0.04 |
ENSDART00000136024
|
ftr14l
|
finTRIM family, member 14-like |
| chr4_+_47751483 | 0.04 |
ENSDART00000160043
|
si:ch211-196f19.1
|
si:ch211-196f19.1 |
| chr19_+_10847118 | 0.04 |
ENSDART00000189784
|
apoa4a
|
apolipoprotein A-IV a |
| chr13_+_25434932 | 0.04 |
ENSDART00000128562
|
si:dkey-51a16.9
|
si:dkey-51a16.9 |
| chr21_-_26918901 | 0.04 |
ENSDART00000100685
|
lrfn4a
|
leucine rich repeat and fibronectin type III domain containing 4a |
| chr23_+_35650771 | 0.03 |
ENSDART00000005158
|
ccnt1
|
cyclin T1 |
| chr17_-_49443399 | 0.03 |
ENSDART00000155293
|
cga
|
glycoprotein hormones, alpha polypeptide |
| chr15_-_33640890 | 0.03 |
ENSDART00000167705
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr17_-_2817035 | 0.03 |
ENSDART00000152002
|
gpr65
|
G protein-coupled receptor 65 |
| chr22_+_25672155 | 0.03 |
ENSDART00000087769
|
si:ch211-250e5.2
|
si:ch211-250e5.2 |
| chr15_+_46357080 | 0.03 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr22_+_2504524 | 0.03 |
ENSDART00000136686
|
znf1170
|
zinc finger protein 1170 |
| chr16_-_11798994 | 0.03 |
ENSDART00000135408
|
cnfn
|
cornifelin |
| chr5_-_35953472 | 0.02 |
ENSDART00000143448
|
rxfp2l
|
relaxin/insulin-like family peptide receptor 2, like |
| chr4_+_5848229 | 0.02 |
ENSDART00000161101
ENSDART00000067357 |
lyrm5a
|
LYR motif containing 5a |
| chr19_-_5265155 | 0.02 |
ENSDART00000145003
|
prf1.3
|
perforin 1.3 |
| chr16_-_21038015 | 0.02 |
ENSDART00000059239
|
snx10b
|
sorting nexin 10b |
| chr2_+_6243144 | 0.02 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr3_-_21288202 | 0.02 |
ENSDART00000191766
ENSDART00000187319 |
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr15_+_24704798 | 0.02 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr1_+_45121393 | 0.02 |
ENSDART00000142702
|
muc13a
|
mucin 13a, cell surface associated |
| chr21_+_25231160 | 0.02 |
ENSDART00000063089
ENSDART00000139127 |
gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr6_-_54107269 | 0.02 |
ENSDART00000190017
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr2_-_27319914 | 0.02 |
ENSDART00000182011
|
dsela
|
dermatan sulfate epimerase-like a |
| chr9_-_44953664 | 0.02 |
ENSDART00000188558
ENSDART00000185210 |
vil1
|
villin 1 |
| chr3_-_28250722 | 0.02 |
ENSDART00000165936
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr9_-_22339582 | 0.02 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr18_+_50523870 | 0.01 |
ENSDART00000151593
|
ubl7b
|
ubiquitin-like 7b (bone marrow stromal cell-derived) |
| chr20_-_10288156 | 0.01 |
ENSDART00000064110
|
si:dkey-63b1.1
|
si:dkey-63b1.1 |
| chr15_-_16098531 | 0.01 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr24_-_21404367 | 0.01 |
ENSDART00000152093
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr17_+_12698532 | 0.00 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr25_+_3648497 | 0.00 |
ENSDART00000160017
|
CABZ01058261.1
|
|
| chr1_+_11822 | 0.00 |
ENSDART00000166393
|
cep97
|
centrosomal protein 97 |
| chr25_+_31958911 | 0.00 |
ENSDART00000191394
ENSDART00000090727 ENSDART00000185893 |
duox
|
dual oxidase |
| chr3_+_33341640 | 0.00 |
ENSDART00000186352
|
pyya
|
peptide YYa |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou6f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005991 | trehalose metabolic process(GO:0005991) |
| 0.2 | 1.0 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:0090069 | regulation of ribosome biogenesis(GO:0090069) |
| 0.1 | 0.6 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.1 | 0.4 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.3 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.1 | GO:1903646 | regulation of protein folding(GO:1903332) positive regulation of protein folding(GO:1903334) regulation of chaperone-mediated protein folding(GO:1903644) positive regulation of chaperone-mediated protein folding(GO:1903646) |
| 0.0 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.3 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.0 | 0.2 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0046037 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) regulation of lamellipodium morphogenesis(GO:2000392) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.0 | GO:0060262 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.0 | 0.3 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.0 | GO:0010893 | positive regulation of steroid biosynthetic process(GO:0010893) |
| 0.0 | 0.5 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.3 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.1 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.0 | 0.1 | GO:0072380 | TRC complex(GO:0072380) |
| 0.0 | 0.9 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.4 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.2 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0015927 | alpha,alpha-trehalase activity(GO:0004555) trehalase activity(GO:0015927) |
| 0.0 | 0.4 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.2 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |