Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
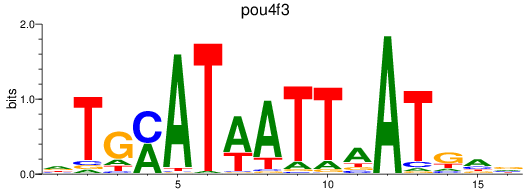
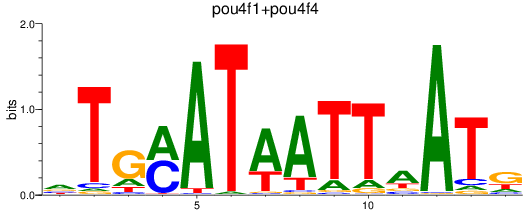
Results for pou4f3_pou4f1+pou4f4
Z-value: 0.46
Transcription factors associated with pou4f3_pou4f1+pou4f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f3
|
ENSDARG00000006206 | POU class 4 homeobox 3 |
|
pou4f1
|
ENSDARG00000005559 | POU class 4 homeobox 1 |
|
pou4f4
|
ENSDARG00000044375 | zgc |
|
pou4f4
|
ENSDARG00000111929 | zgc |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f1 | dr11_v1_chr6_+_4539953_4539953 | -0.48 | 4.4e-02 | Click! |
| zgc:158291 | dr11_v1_chr21_+_38312549_38312549 | 0.19 | 4.6e-01 | Click! |
| pou4f3 | dr11_v1_chr9_+_54290896_54290896 | -0.08 | 7.7e-01 | Click! |
Activity profile of pou4f3_pou4f1+pou4f4 motif
Sorted Z-values of pou4f3_pou4f1+pou4f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_10177442 | 1.44 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr7_+_12371061 | 1.34 |
ENSDART00000053860
|
saxo2
|
stabilizer of axonemal microtubules 2 |
| chr17_-_51893123 | 0.78 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr5_+_58372164 | 0.77 |
ENSDART00000057910
|
nrgna
|
neurogranin (protein kinase C substrate, RC3) a |
| chr15_+_34988148 | 0.68 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr1_-_55270966 | 0.63 |
ENSDART00000152807
|
si:ch211-286b5.5
|
si:ch211-286b5.5 |
| chr10_+_39084354 | 0.57 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr4_+_6869847 | 0.53 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr7_-_28148310 | 0.51 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr20_+_22045089 | 0.45 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr20_-_31075972 | 0.41 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr24_-_4765740 | 0.40 |
ENSDART00000121576
|
cpb1
|
carboxypeptidase B1 (tissue) |
| chr23_+_24085531 | 0.39 |
ENSDART00000139710
|
ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr20_+_21022641 | 0.39 |
ENSDART00000152561
ENSDART00000141591 |
si:dkey-219e20.2
|
si:dkey-219e20.2 |
| chr3_+_26734162 | 0.38 |
ENSDART00000114552
|
si:dkey-202l16.5
|
si:dkey-202l16.5 |
| chr17_-_25649079 | 0.38 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr19_+_41551543 | 0.38 |
ENSDART00000112364
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr23_+_19977120 | 0.36 |
ENSDART00000089342
|
cfap126
|
cilia and flagella associated protein 126 |
| chr20_-_36617313 | 0.35 |
ENSDART00000172395
ENSDART00000152856 |
enah
|
enabled homolog (Drosophila) |
| chr16_+_10318893 | 0.35 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr6_+_13039951 | 0.35 |
ENSDART00000091700
|
catip
|
ciliogenesis associated TTC17 interacting protein |
| chr12_+_22560067 | 0.34 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr23_-_27692717 | 0.34 |
ENSDART00000053878
ENSDART00000145028 |
IKZF4
|
si:dkey-166n8.9 |
| chr2_-_43196595 | 0.32 |
ENSDART00000141087
|
crema
|
cAMP responsive element modulator a |
| chr12_-_35944654 | 0.29 |
ENSDART00000162579
ENSDART00000164199 |
dnai2a
|
dynein, axonemal, intermediate chain 2a |
| chr12_-_19151708 | 0.29 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr22_-_29906764 | 0.29 |
ENSDART00000019786
|
smc3
|
structural maintenance of chromosomes 3 |
| chr15_-_23206562 | 0.29 |
ENSDART00000110360
|
ccdc153
|
coiled-coil domain containing 153 |
| chr9_-_33081978 | 0.28 |
ENSDART00000100918
|
zgc:172053
|
zgc:172053 |
| chr22_-_3564563 | 0.27 |
ENSDART00000145114
|
ptprsa
|
protein tyrosine phosphatase, receptor type, s, a |
| chr3_+_1107102 | 0.27 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr25_+_34915576 | 0.26 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr5_+_42393896 | 0.26 |
ENSDART00000189550
|
BX548073.13
|
|
| chr5_-_54672763 | 0.25 |
ENSDART00000159009
|
spag8
|
sperm associated antigen 8 |
| chr19_+_41551335 | 0.25 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr7_+_73397283 | 0.25 |
ENSDART00000174390
|
CABZ01081780.1
|
|
| chr25_+_3358701 | 0.24 |
ENSDART00000104877
|
chchd3b
|
coiled-coil-helix-coiled-coil-helix domain containing 3b |
| chr11_+_6819050 | 0.24 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr13_+_27298673 | 0.24 |
ENSDART00000142922
|
slc17a5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr12_+_2446837 | 0.23 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr9_+_1039215 | 0.23 |
ENSDART00000192186
|
DOCK9 (1 of many)
|
dedicator of cytokinesis 9 |
| chr6_+_21740672 | 0.23 |
ENSDART00000193734
|
lhfpl4a
|
lipoma HMGIC fusion partner-like 4a |
| chr15_-_25083200 | 0.23 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr17_+_2162916 | 0.21 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr3_+_36313532 | 0.21 |
ENSDART00000151305
|
slc16a6b
|
solute carrier family 16, member 6b |
| chr25_+_34915762 | 0.20 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr6_-_16394528 | 0.20 |
ENSDART00000089445
|
agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr16_+_43401005 | 0.20 |
ENSDART00000110994
|
sqlea
|
squalene epoxidase a |
| chr14_+_6991142 | 0.19 |
ENSDART00000157635
ENSDART00000059918 |
hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chr7_+_27059330 | 0.19 |
ENSDART00000173919
|
plekha7b
|
pleckstrin homology domain containing, family A member 7b |
| chr13_+_27951688 | 0.18 |
ENSDART00000050303
|
b3gat2
|
beta-1,3-glucuronyltransferase 2 (glucuronosyltransferase S) |
| chr17_-_18797245 | 0.18 |
ENSDART00000045991
|
vrk1
|
vaccinia related kinase 1 |
| chr9_+_38481780 | 0.18 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr3_-_23407720 | 0.18 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr16_-_24605969 | 0.18 |
ENSDART00000163305
ENSDART00000167121 |
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr17_+_26352372 | 0.17 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr8_+_30600386 | 0.17 |
ENSDART00000164976
|
specc1la
|
sperm antigen with calponin homology and coiled-coil domains 1-like a |
| chr10_+_15511885 | 0.17 |
ENSDART00000165663
|
erbin
|
erbb2 interacting protein |
| chr4_+_21867522 | 0.16 |
ENSDART00000140400
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr25_-_22187397 | 0.16 |
ENSDART00000123211
ENSDART00000139110 |
pkp3a
|
plakophilin 3a |
| chr23_+_20705849 | 0.16 |
ENSDART00000079538
|
ccdc30
|
coiled-coil domain containing 30 |
| chr17_+_2727807 | 0.16 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr10_-_31015535 | 0.15 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr12_-_10705916 | 0.15 |
ENSDART00000164038
|
FO704786.1
|
|
| chr11_+_37137196 | 0.15 |
ENSDART00000172950
|
wnk2
|
WNK lysine deficient protein kinase 2 |
| chr11_+_25101220 | 0.15 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr20_+_37661547 | 0.15 |
ENSDART00000007253
|
aig1
|
androgen-induced 1 (H. sapiens) |
| chr23_+_23658474 | 0.15 |
ENSDART00000162838
|
agrn
|
agrin |
| chr9_-_7652792 | 0.14 |
ENSDART00000137957
|
dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr21_-_13123176 | 0.14 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr9_+_7030016 | 0.14 |
ENSDART00000148047
ENSDART00000148181 |
inpp4aa
|
inositol polyphosphate-4-phosphatase type I Aa |
| chr13_+_45524475 | 0.14 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr13_+_4409294 | 0.14 |
ENSDART00000146437
|
si:ch211-130h14.4
|
si:ch211-130h14.4 |
| chr20_+_17783404 | 0.13 |
ENSDART00000181946
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr25_-_13614702 | 0.13 |
ENSDART00000165510
ENSDART00000190959 |
fa2h
|
fatty acid 2-hydroxylase |
| chr17_+_26208630 | 0.12 |
ENSDART00000087084
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr23_+_20689255 | 0.12 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr11_+_44356707 | 0.12 |
ENSDART00000165219
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr11_+_44356504 | 0.12 |
ENSDART00000160678
|
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr25_-_13614863 | 0.12 |
ENSDART00000121859
|
fa2h
|
fatty acid 2-hydroxylase |
| chr22_+_13886821 | 0.12 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr10_-_27566481 | 0.11 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr25_+_8407892 | 0.11 |
ENSDART00000153536
|
muc5.2
|
mucin 5.2 |
| chr5_+_28313824 | 0.11 |
ENSDART00000135878
|
si:dkeyp-86b9.1
|
si:dkeyp-86b9.1 |
| chr9_+_34319333 | 0.11 |
ENSDART00000035522
ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr4_+_9669717 | 0.11 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr19_-_10196370 | 0.11 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr9_-_68934 | 0.11 |
ENSDART00000054594
ENSDART00000009389 |
il10rb
|
interleukin 10 receptor, beta |
| chr20_+_20638034 | 0.10 |
ENSDART00000189759
|
rtn1b
|
reticulon 1b |
| chr3_-_23406964 | 0.10 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr10_-_29892486 | 0.10 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr20_+_41756996 | 0.10 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr6_+_3280939 | 0.10 |
ENSDART00000151359
|
kdm4aa
|
lysine (K)-specific demethylase 4A, genome duplicate a |
| chr7_+_56577906 | 0.10 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr7_+_20518218 | 0.10 |
ENSDART00000052916
|
zgc:103586
|
zgc:103586 |
| chr19_+_37620342 | 0.10 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr13_+_25286538 | 0.10 |
ENSDART00000180282
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr10_-_10018120 | 0.10 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr10_+_10351685 | 0.10 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr13_-_32635859 | 0.10 |
ENSDART00000146249
ENSDART00000145395 ENSDART00000148040 ENSDART00000100650 |
matn3b
|
matrilin 3b |
| chr9_+_44431174 | 0.10 |
ENSDART00000149726
|
ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr7_-_49892991 | 0.10 |
ENSDART00000126240
|
cd44a
|
CD44 molecule (Indian blood group) a |
| chr20_+_27298783 | 0.10 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr2_+_41926707 | 0.09 |
ENSDART00000023208
|
zgc:110183
|
zgc:110183 |
| chr6_-_35446110 | 0.09 |
ENSDART00000058773
|
rgs16
|
regulator of G protein signaling 16 |
| chr24_+_36840652 | 0.09 |
ENSDART00000088168
|
CABZ01055347.1
|
|
| chr6_-_8295657 | 0.09 |
ENSDART00000185135
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr2_+_22702488 | 0.09 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr4_-_22338906 | 0.09 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr14_-_36799280 | 0.09 |
ENSDART00000168615
|
rnf130
|
ring finger protein 130 |
| chr24_-_3419998 | 0.09 |
ENSDART00000066839
|
slc35g2b
|
solute carrier family 35, member G2b |
| chr2_+_22042745 | 0.09 |
ENSDART00000132039
|
tox
|
thymocyte selection-associated high mobility group box |
| chr1_+_25783801 | 0.08 |
ENSDART00000102455
|
gucy1a1
|
guanylate cyclase 1 soluble subunit alpha 1 |
| chr10_+_42423318 | 0.08 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr20_-_2361226 | 0.08 |
ENSDART00000172130
|
EPB41L2
|
si:ch73-18b11.1 |
| chr11_+_34523132 | 0.08 |
ENSDART00000192257
|
zmat3
|
zinc finger, matrin-type 3 |
| chr13_+_31757331 | 0.08 |
ENSDART00000044282
|
hif1aa
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) a |
| chr16_+_14029283 | 0.08 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr24_+_14713776 | 0.08 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr12_-_7639120 | 0.08 |
ENSDART00000126712
ENSDART00000126219 |
ccdc6b
|
coiled-coil domain containing 6b |
| chr3_-_29968015 | 0.08 |
ENSDART00000077119
ENSDART00000139310 |
bcat2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr8_+_7975745 | 0.07 |
ENSDART00000137920
|
si:ch211-169p10.1
|
si:ch211-169p10.1 |
| chr10_+_40321067 | 0.07 |
ENSDART00000109816
|
gltpb
|
glycolipid transfer protein b |
| chr13_-_21688176 | 0.07 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr21_+_170038 | 0.07 |
ENSDART00000157614
|
klhl8
|
kelch-like family member 8 |
| chr14_-_47849216 | 0.07 |
ENSDART00000192796
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr21_+_26389391 | 0.07 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr22_-_32360464 | 0.07 |
ENSDART00000148886
|
pcbp4
|
poly(rC) binding protein 4 |
| chr16_+_16849220 | 0.07 |
ENSDART00000047409
ENSDART00000142155 |
myh14
|
myosin, heavy chain 14, non-muscle |
| chr25_+_21833287 | 0.07 |
ENSDART00000187606
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr2_-_24348642 | 0.07 |
ENSDART00000181739
|
ano8a
|
anoctamin 8a |
| chr4_-_5775507 | 0.07 |
ENSDART00000021753
|
ccnc
|
cyclin C |
| chr18_-_40884087 | 0.06 |
ENSDART00000059194
|
snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr21_-_13972745 | 0.06 |
ENSDART00000143874
|
akna
|
AT-hook transcription factor |
| chr2_-_24348948 | 0.06 |
ENSDART00000136559
|
ano8a
|
anoctamin 8a |
| chr2_-_45510699 | 0.06 |
ENSDART00000024034
ENSDART00000145634 |
gpsm2
|
G protein signaling modulator 2 |
| chr16_-_9802449 | 0.06 |
ENSDART00000081208
|
tapbpl
|
TAP binding protein (tapasin)-like |
| chr21_-_26691959 | 0.06 |
ENSDART00000149702
ENSDART00000149840 |
bscl2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr3_-_16055432 | 0.06 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr22_-_17585618 | 0.06 |
ENSDART00000183123
|
pip5k1ca
|
phosphatidylinositol-4-phosphate 5-kinase, type I, gamma a |
| chr20_-_6196989 | 0.06 |
ENSDART00000013343
|
b4galt6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr24_+_17142881 | 0.06 |
ENSDART00000177272
ENSDART00000192259 ENSDART00000191029 |
mllt10
|
MLLT10, histone lysine methyltransferase DOT1L cofactor |
| chr17_+_33453689 | 0.06 |
ENSDART00000156894
|
rin3
|
Ras and Rab interactor 3 |
| chr11_+_2416064 | 0.05 |
ENSDART00000067117
|
ube2v1
|
ubiquitin-conjugating enzyme E2 variant 1 |
| chr20_-_7293837 | 0.05 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr20_+_9355912 | 0.05 |
ENSDART00000142337
|
si:dkey-174m14.3
|
si:dkey-174m14.3 |
| chr3_-_28665291 | 0.05 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr5_-_29195063 | 0.05 |
ENSDART00000109926
|
man1b1b
|
mannosidase, alpha, class 1B, member 1b |
| chr15_+_24704798 | 0.05 |
ENSDART00000192470
|
LRRC75A
|
si:dkey-151p21.7 |
| chr11_+_36409457 | 0.05 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr2_+_38742338 | 0.05 |
ENSDART00000177290
|
carmil3
|
capping protein regulator and myosin 1 linker 3 |
| chr3_+_23731109 | 0.05 |
ENSDART00000131410
|
hoxb3a
|
homeobox B3a |
| chr7_-_25895189 | 0.05 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr19_-_3574060 | 0.05 |
ENSDART00000105120
|
tmem170b
|
transmembrane protein 170B |
| chr22_+_5687615 | 0.05 |
ENSDART00000133241
ENSDART00000019854 ENSDART00000138102 |
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr22_+_16535575 | 0.05 |
ENSDART00000083063
|
tal1
|
T-cell acute lymphocytic leukemia 1 |
| chr10_+_44824072 | 0.05 |
ENSDART00000162669
|
slc4a5
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 5 |
| chr8_+_17775247 | 0.05 |
ENSDART00000112356
|
si:ch211-150o23.3
|
si:ch211-150o23.3 |
| chr16_-_46660680 | 0.04 |
ENSDART00000159209
ENSDART00000191929 |
tmem176l.4
|
transmembrane protein 176l.4 |
| chr23_+_11285662 | 0.04 |
ENSDART00000111028
|
chl1a
|
cell adhesion molecule L1-like a |
| chr1_-_19502322 | 0.04 |
ENSDART00000181888
ENSDART00000044030 |
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr25_+_19201231 | 0.04 |
ENSDART00000067323
|
hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr2_-_31651297 | 0.04 |
ENSDART00000132955
|
si:ch211-106h4.9
|
si:ch211-106h4.9 |
| chr10_+_19528321 | 0.04 |
ENSDART00000184816
|
vsig8a
|
V-set and immunoglobulin domain containing 8a |
| chr3_-_61336841 | 0.04 |
ENSDART00000155414
|
tecpr1b
|
tectonin beta-propeller repeat containing 1b |
| chr18_-_23875370 | 0.04 |
ENSDART00000130163
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_48171112 | 0.04 |
ENSDART00000156258
|
pfkpb
|
phosphofructokinase, platelet b |
| chr15_+_45586471 | 0.04 |
ENSDART00000165699
|
cldn15lb
|
claudin 15-like b |
| chr13_-_32899322 | 0.04 |
ENSDART00000133882
|
rock2a
|
rho-associated, coiled-coil containing protein kinase 2a |
| chr15_-_14469704 | 0.04 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr17_+_25444605 | 0.04 |
ENSDART00000149804
|
clic4
|
chloride intracellular channel 4 |
| chr5_-_68058168 | 0.04 |
ENSDART00000177026
|
rnf167
|
ring finger protein 167 |
| chr16_-_55259199 | 0.04 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr6_-_41229787 | 0.04 |
ENSDART00000065013
|
synpr
|
synaptoporin |
| chr2_+_39108339 | 0.04 |
ENSDART00000085675
|
clstn2
|
calsyntenin 2 |
| chr22_+_10651726 | 0.04 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr4_+_21866851 | 0.04 |
ENSDART00000177640
|
acss3
|
acyl-CoA synthetase short chain family member 3 |
| chr25_-_15040369 | 0.04 |
ENSDART00000159342
ENSDART00000166490 |
pax6a
|
paired box 6a |
| chr7_-_69298271 | 0.04 |
ENSDART00000122935
|
phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr12_-_44072716 | 0.03 |
ENSDART00000162509
|
si:ch73-329n5.3
|
si:ch73-329n5.3 |
| chr12_+_8168272 | 0.03 |
ENSDART00000054092
|
arid5b
|
AT-rich interaction domain 5B |
| chr7_-_17690756 | 0.03 |
ENSDART00000173759
|
map4k2
|
mitogen-activated protein kinase kinase kinase kinase 2 |
| chr4_+_20063279 | 0.03 |
ENSDART00000024925
|
gcc1
|
GRIP and coiled-coil domain containing 1 |
| chr9_-_25989989 | 0.03 |
ENSDART00000090052
|
arhgap15
|
Rho GTPase activating protein 15 |
| chr1_+_44491077 | 0.03 |
ENSDART00000073736
|
rtn4rl2a
|
reticulon 4 receptor-like 2 a |
| chr12_+_4260779 | 0.03 |
ENSDART00000081382
|
mmp25b
|
matrix metallopeptidase 25b |
| chr1_-_22861348 | 0.03 |
ENSDART00000139412
|
SMIM18
|
si:dkey-92j12.6 |
| chr22_-_23748284 | 0.03 |
ENSDART00000162005
|
cfhl2
|
complement factor H like 2 |
| chr11_+_30057762 | 0.03 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr6_+_4299164 | 0.03 |
ENSDART00000159759
|
nbeal1
|
neurobeachin-like 1 |
| chr11_-_38513978 | 0.03 |
ENSDART00000123381
|
si:ch211-117k10.3
|
si:ch211-117k10.3 |
| chr25_+_14507567 | 0.03 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr19_+_30662529 | 0.03 |
ENSDART00000175662
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr7_+_27317174 | 0.03 |
ENSDART00000193058
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr8_-_27858458 | 0.03 |
ENSDART00000132632
ENSDART00000136562 |
cttnbp2nlb
|
CTTNBP2 N-terminal like b |
| chr5_+_12888260 | 0.02 |
ENSDART00000175916
|
gal3st1a
|
galactose-3-O-sulfotransferase 1a |
| chr10_+_38775408 | 0.02 |
ENSDART00000125045
|
dscama
|
Down syndrome cell adhesion molecule a |
| chr17_+_9009098 | 0.02 |
ENSDART00000180856
|
akap6
|
A kinase (PRKA) anchor protein 6 |
| chr6_+_27418541 | 0.02 |
ENSDART00000187410
|
crocc2
|
ciliary rootlet coiled-coil, rootletin family member 2 |
| chr13_+_36622100 | 0.02 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f3_pou4f1+pou4f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.2 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.1 | 1.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.0 | 0.2 | GO:0048313 | organelle inheritance(GO:0048308) Golgi inheritance(GO:0048313) |
| 0.0 | 0.1 | GO:0035142 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.0 | 0.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:0010885 | regulation of cholesterol storage(GO:0010885) |
| 0.0 | 0.8 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.0 | 0.3 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.3 | GO:0099560 | synaptic membrane adhesion(GO:0099560) |
| 0.0 | 0.1 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 0.0 | 2.1 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.0 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.0 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.0 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.6 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.3 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0080132 | fatty acid alpha-hydroxylase activity(GO:0080132) |
| 0.1 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.2 | GO:0050218 | propionate-CoA ligase activity(GO:0050218) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.4 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0035620 | ceramide transporter activity(GO:0035620) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.3 | GO:0034062 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |