Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
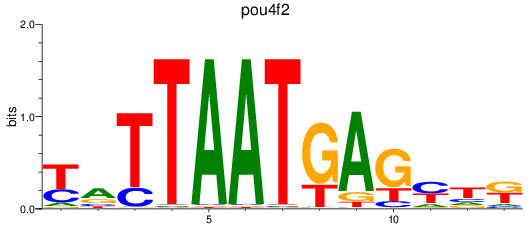
Results for pou4f2
Z-value: 0.28
Transcription factors associated with pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou4f2
|
ENSDARG00000069737 | POU class 4 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou4f2 | dr11_v1_chr1_+_36436936_36436936 | 0.32 | 1.9e-01 | Click! |
Activity profile of pou4f2 motif
Sorted Z-values of pou4f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_24952902 | 0.56 |
ENSDART00000189086
ENSDART00000014868 |
calm3a
|
calmodulin 3a (phosphorylase kinase, delta) |
| chr25_-_16581233 | 0.46 |
ENSDART00000155312
|
smc1b
|
structural maintenance of chromosomes 1B |
| chr21_+_36623162 | 0.37 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr1_-_55270966 | 0.37 |
ENSDART00000152807
|
si:ch211-286b5.5
|
si:ch211-286b5.5 |
| chr4_-_1801519 | 0.31 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr5_+_42381273 | 0.29 |
ENSDART00000142387
|
pimr58
|
Pim proto-oncogene, serine/threonine kinase, related 58 |
| chr24_-_21923930 | 0.28 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr13_-_42400647 | 0.25 |
ENSDART00000043069
|
march5
|
membrane-associated ring finger (C3HC4) 5 |
| chr11_-_2478374 | 0.25 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr10_-_35103208 | 0.25 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr5_-_38155005 | 0.24 |
ENSDART00000097770
|
gucy2d
|
guanylate cyclase 2D, retinal |
| chr7_+_73630751 | 0.23 |
ENSDART00000159745
|
PCP4L1
|
si:dkey-46i9.1 |
| chr17_+_15433518 | 0.23 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr12_-_35830625 | 0.23 |
ENSDART00000180028
|
CU459056.1
|
|
| chr19_-_31402429 | 0.22 |
ENSDART00000137292
|
tmem106bb
|
transmembrane protein 106Bb |
| chr2_-_40135942 | 0.22 |
ENSDART00000176951
ENSDART00000098632 ENSDART00000148563 ENSDART00000149895 |
epha4a
|
eph receptor A4a |
| chr17_+_15433671 | 0.21 |
ENSDART00000149568
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr7_+_32369026 | 0.20 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr1_-_17715493 | 0.20 |
ENSDART00000133027
|
si:dkey-256e7.8
|
si:dkey-256e7.8 |
| chr8_+_52314542 | 0.19 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr23_-_28141419 | 0.17 |
ENSDART00000133039
|
tac3a
|
tachykinin 3a |
| chr8_+_17167876 | 0.17 |
ENSDART00000134665
|
cenph
|
centromere protein H |
| chr10_+_39084354 | 0.17 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr12_-_10409961 | 0.16 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr24_+_26432541 | 0.16 |
ENSDART00000105784
ENSDART00000122554 |
cldn11b
|
claudin 11b |
| chr19_+_1688727 | 0.15 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr18_+_2228737 | 0.15 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr8_-_13454281 | 0.15 |
ENSDART00000141959
|
CR354547.2
|
|
| chr22_-_13042992 | 0.15 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr25_-_13842618 | 0.15 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr25_+_6186823 | 0.15 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr8_+_44714336 | 0.14 |
ENSDART00000145801
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr4_+_11695979 | 0.14 |
ENSDART00000137736
|
mkln1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr16_+_11242443 | 0.13 |
ENSDART00000024935
|
gsk3ab
|
glycogen synthase kinase 3 alpha b |
| chr6_+_54187643 | 0.13 |
ENSDART00000056830
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr20_+_41756996 | 0.12 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr1_+_144284 | 0.12 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr13_-_33194647 | 0.12 |
ENSDART00000134383
|
trip11
|
thyroid hormone receptor interactor 11 |
| chr5_+_51443009 | 0.11 |
ENSDART00000083350
|
rasgrf2b
|
Ras protein-specific guanine nucleotide-releasing factor 2b |
| chr1_+_10318089 | 0.11 |
ENSDART00000029774
|
pip4p1b
|
phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1b |
| chr8_+_8927870 | 0.11 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr8_+_16025554 | 0.11 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr7_+_19552381 | 0.11 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr16_-_20932896 | 0.11 |
ENSDART00000180646
|
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr4_-_1360495 | 0.10 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr7_-_7764287 | 0.10 |
ENSDART00000173021
ENSDART00000113131 |
intu
|
inturned planar cell polarity protein |
| chr5_+_44064764 | 0.10 |
ENSDART00000143843
|
si:dkey-84j12.1
|
si:dkey-84j12.1 |
| chr18_-_48550426 | 0.09 |
ENSDART00000145189
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr6_+_39184236 | 0.09 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr11_-_45138857 | 0.09 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr5_+_17624463 | 0.09 |
ENSDART00000183869
ENSDART00000081064 |
fbrsl1
|
fibrosin-like 1 |
| chr3_+_45687266 | 0.09 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr6_+_27146671 | 0.09 |
ENSDART00000156792
|
kif1aa
|
kinesin family member 1Aa |
| chr16_-_12173554 | 0.09 |
ENSDART00000110567
ENSDART00000155935 |
clstn3
|
calsyntenin 3 |
| chr18_-_15551360 | 0.09 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr5_-_72415578 | 0.09 |
ENSDART00000029014
|
pax8
|
paired box 8 |
| chr11_-_3860534 | 0.09 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr3_-_61362398 | 0.09 |
ENSDART00000156177
|
si:dkey-111k8.3
|
si:dkey-111k8.3 |
| chr14_+_45883687 | 0.09 |
ENSDART00000114790
|
flrt1b
|
fibronectin leucine rich transmembrane protein 1b |
| chr11_-_3860199 | 0.08 |
ENSDART00000082420
|
gata2a
|
GATA binding protein 2a |
| chr17_-_49438873 | 0.08 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr1_-_50859053 | 0.08 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr1_+_25801648 | 0.08 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr18_-_1185772 | 0.08 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr3_-_23406964 | 0.08 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr16_+_29303971 | 0.08 |
ENSDART00000087149
|
hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr23_+_41912151 | 0.08 |
ENSDART00000191115
|
podn
|
podocan |
| chr22_-_347424 | 0.08 |
ENSDART00000067633
|
necap2
|
NECAP endocytosis associated 2 |
| chr17_+_11675362 | 0.07 |
ENSDART00000157911
|
kif26ba
|
kinesin family member 26Ba |
| chr14_-_7306983 | 0.07 |
ENSDART00000158914
|
si:ch211-51f19.1
|
si:ch211-51f19.1 |
| chr23_+_1276006 | 0.07 |
ENSDART00000162294
|
utrn
|
utrophin |
| chr13_+_35339182 | 0.07 |
ENSDART00000019323
|
jag1b
|
jagged 1b |
| chr16_-_12173399 | 0.07 |
ENSDART00000142574
|
clstn3
|
calsyntenin 3 |
| chr14_+_28486213 | 0.07 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr21_+_43328685 | 0.07 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr21_-_7035599 | 0.07 |
ENSDART00000139777
|
si:ch211-93g21.1
|
si:ch211-93g21.1 |
| chr20_-_52705325 | 0.06 |
ENSDART00000136807
ENSDART00000136126 |
si:ch211-221n20.7
|
si:ch211-221n20.7 |
| chr8_+_17884569 | 0.06 |
ENSDART00000134660
|
slc44a5b
|
solute carrier family 44, member 5b |
| chr9_-_23944470 | 0.06 |
ENSDART00000138754
|
col6a3
|
collagen, type VI, alpha 3 |
| chr11_+_1575435 | 0.06 |
ENSDART00000155713
ENSDART00000156562 |
si:dkey-40c23.1
|
si:dkey-40c23.1 |
| chr19_+_9459050 | 0.06 |
ENSDART00000186419
|
si:ch211-288g17.3
|
si:ch211-288g17.3 |
| chr17_+_26965351 | 0.05 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr9_+_21793565 | 0.05 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr11_-_31226578 | 0.05 |
ENSDART00000109698
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr11_-_37509001 | 0.05 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr2_-_21082695 | 0.05 |
ENSDART00000032502
|
nebl
|
nebulette |
| chr21_-_20733615 | 0.05 |
ENSDART00000145544
|
si:ch211-22d5.2
|
si:ch211-22d5.2 |
| chr18_-_2433011 | 0.05 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr8_+_17069577 | 0.04 |
ENSDART00000131847
|
si:dkey-190g11.3
|
si:dkey-190g11.3 |
| chr4_-_9836342 | 0.04 |
ENSDART00000150519
|
ttc41
|
tetratricopeptide repeat domain 41 |
| chr17_+_8183393 | 0.04 |
ENSDART00000155957
|
tulp4b
|
tubby like protein 4b |
| chr17_+_28706946 | 0.04 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr13_+_36622100 | 0.04 |
ENSDART00000133198
|
si:ch211-67f24.7
|
si:ch211-67f24.7 |
| chr4_+_2637947 | 0.04 |
ENSDART00000130623
|
dus4l
|
dihydrouridine synthase 4-like (S. cerevisiae) |
| chr21_-_40348790 | 0.04 |
ENSDART00000178123
|
CR847523.1
|
|
| chr14_-_31465905 | 0.04 |
ENSDART00000173108
|
gpc3
|
glypican 3 |
| chr10_+_36178713 | 0.03 |
ENSDART00000140816
|
or108-3
|
odorant receptor, family D, subfamily 108, member 3 |
| chr11_+_44502410 | 0.03 |
ENSDART00000172998
|
ero1b
|
endoplasmic reticulum oxidoreductase beta |
| chr7_+_25915089 | 0.03 |
ENSDART00000173648
|
hmgb3a
|
high mobility group box 3a |
| chr16_-_26074529 | 0.03 |
ENSDART00000148653
ENSDART00000148923 |
tmem145
|
transmembrane protein 145 |
| chr3_-_58798377 | 0.03 |
ENSDART00000161248
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr11_-_42918971 | 0.03 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr20_+_18325556 | 0.03 |
ENSDART00000123559
|
znf521
|
zinc finger protein 521 |
| chr13_+_22476742 | 0.03 |
ENSDART00000078759
ENSDART00000130101 ENSDART00000137220 ENSDART00000133065 ENSDART00000147348 |
ldb3a
|
LIM domain binding 3a |
| chr15_-_41744509 | 0.03 |
ENSDART00000176952
ENSDART00000156690 |
ftr88
|
finTRIM family, member 88 |
| chr17_+_26352372 | 0.03 |
ENSDART00000155177
|
grid1a
|
glutamate receptor, ionotropic, delta 1a |
| chr18_-_40509006 | 0.03 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr3_-_58798815 | 0.02 |
ENSDART00000082920
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr7_+_25059845 | 0.02 |
ENSDART00000077215
|
ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr4_-_28335184 | 0.02 |
ENSDART00000178149
ENSDART00000043737 |
celsr1a
|
cadherin EGF LAG seven-pass G-type receptor 1a |
| chr3_+_56276346 | 0.02 |
ENSDART00000157770
|
PRKCA
|
si:ch73-374l24.1 |
| chr19_-_32641725 | 0.02 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr14_+_46287296 | 0.02 |
ENSDART00000183620
|
cabp2b
|
calcium binding protein 2b |
| chr19_+_42470396 | 0.02 |
ENSDART00000191679
|
si:dkey-166k12.1
|
si:dkey-166k12.1 |
| chr12_-_35787801 | 0.02 |
ENSDART00000171682
|
aatkb
|
apoptosis-associated tyrosine kinase b |
| chr6_+_4033832 | 0.02 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr1_-_38195012 | 0.02 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr14_-_32959851 | 0.02 |
ENSDART00000075157
|
chic1
|
cysteine-rich hydrophobic domain 1 |
| chr21_+_28958471 | 0.02 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr10_-_17159761 | 0.01 |
ENSDART00000080449
|
slc5a1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr11_+_3585934 | 0.01 |
ENSDART00000055694
|
cdab
|
cytidine deaminase b |
| chr11_+_38280454 | 0.01 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr2_+_36939749 | 0.01 |
ENSDART00000005382
|
gadd45bb
|
growth arrest and DNA-damage-inducible, beta b |
| chr9_-_32753535 | 0.01 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr13_+_31180084 | 0.01 |
ENSDART00000133774
|
ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr5_-_68782641 | 0.01 |
ENSDART00000141699
|
mepce
|
methylphosphate capping enzyme |
| chr5_-_63515210 | 0.01 |
ENSDART00000022348
|
prdm12b
|
PR domain containing 12b |
| chr3_-_32169754 | 0.00 |
ENSDART00000179010
|
tnnt1
|
troponin T type 1 (skeletal, slow) |
| chr3_+_33341640 | 0.00 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr7_-_33960170 | 0.00 |
ENSDART00000180766
|
skor1a
|
SKI family transcriptional corepressor 1a |
| chr9_+_29430432 | 0.00 |
ENSDART00000125632
|
uggt2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr24_-_40700596 | 0.00 |
ENSDART00000162635
|
smyhc2
|
slow myosin heavy chain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou4f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.2 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.3 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.0 | 0.2 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.2 | GO:0003404 | optic vesicle morphogenesis(GO:0003404) |
| 0.0 | 0.2 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.0 | 0.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.5 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.1 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.2 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.2 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.2 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.4 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.4 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |