Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
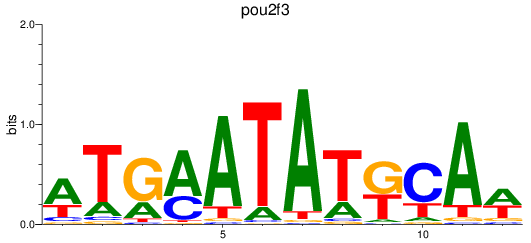
Results for pou2f3
Z-value: 1.25
Transcription factors associated with pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
pou2f3
|
ENSDARG00000052387 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| pou2f3 | dr11_v1_chr5_+_58550291_58550291 | 0.69 | 1.6e-03 | Click! |
Activity profile of pou2f3 motif
Sorted Z-values of pou2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_38049130 | 2.97 |
ENSDART00000097533
|
wwtr1
|
WW domain containing transcription regulator 1 |
| chr5_-_6567464 | 2.38 |
ENSDART00000184985
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr21_+_36623162 | 2.36 |
ENSDART00000027459
|
grk6
|
G protein-coupled receptor kinase 6 |
| chr10_+_39084354 | 2.14 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr17_-_6451801 | 2.10 |
ENSDART00000064700
|
fuca2
|
alpha-L-fucosidase 2 |
| chr25_+_3058924 | 1.96 |
ENSDART00000029580
|
fth1b
|
ferritin, heavy polypeptide 1b |
| chr7_+_13582256 | 1.65 |
ENSDART00000158477
|
ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr21_-_37733571 | 1.59 |
ENSDART00000176214
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr18_-_29962234 | 1.54 |
ENSDART00000144996
|
si:ch73-103b9.2
|
si:ch73-103b9.2 |
| chr7_-_1504382 | 1.52 |
ENSDART00000172770
|
si:zfos-405g10.4
|
si:zfos-405g10.4 |
| chr18_-_20869175 | 1.51 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr21_+_42717424 | 1.34 |
ENSDART00000166936
ENSDART00000172135 |
sh3pxd2b
|
SH3 and PX domains 2B |
| chr2_+_36015049 | 1.33 |
ENSDART00000158276
|
lamc2
|
laminin, gamma 2 |
| chr24_+_34970680 | 1.28 |
ENSDART00000113014
|
rb1cc1
|
RB1-inducible coiled-coil 1 |
| chr6_-_37469775 | 1.25 |
ENSDART00000156546
|
plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr11_-_41132296 | 1.21 |
ENSDART00000162944
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr4_-_1801519 | 1.19 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr8_+_52314542 | 1.12 |
ENSDART00000013059
ENSDART00000125241 |
dbnlb
|
drebrin-like b |
| chr9_-_1702648 | 1.11 |
ENSDART00000102934
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr19_+_1688727 | 1.08 |
ENSDART00000115136
ENSDART00000166744 |
dennd3a
|
DENN/MADD domain containing 3a |
| chr1_+_25801648 | 1.07 |
ENSDART00000129471
|
gucy1b1
|
guanylate cyclase 1 soluble subunit beta 1 |
| chr9_-_8979154 | 1.07 |
ENSDART00000145266
|
ing1
|
inhibitor of growth family, member 1 |
| chr2_+_15048410 | 1.07 |
ENSDART00000058484
|
cnn3b
|
calponin 3, acidic b |
| chr16_+_2820340 | 1.06 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr16_+_23961276 | 1.06 |
ENSDART00000192754
|
apoeb
|
apolipoprotein Eb |
| chr4_+_5341592 | 1.05 |
ENSDART00000123375
ENSDART00000067371 |
zgc:113263
|
zgc:113263 |
| chr19_+_43579786 | 1.04 |
ENSDART00000138404
|
si:ch211-199g17.2
|
si:ch211-199g17.2 |
| chr17_+_12075805 | 1.03 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr9_-_8979468 | 1.02 |
ENSDART00000134646
|
ing1
|
inhibitor of growth family, member 1 |
| chr14_-_34513103 | 0.98 |
ENSDART00000136306
|
zgc:194246
|
zgc:194246 |
| chr11_-_2478374 | 0.98 |
ENSDART00000173205
|
si:ch73-267c23.10
|
si:ch73-267c23.10 |
| chr10_+_26667475 | 0.97 |
ENSDART00000133281
ENSDART00000147013 |
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr17_-_15667198 | 0.96 |
ENSDART00000142972
ENSDART00000132571 ENSDART00000189936 |
manea
|
mannosidase, endo-alpha |
| chr20_-_46817223 | 0.95 |
ENSDART00000100336
|
esrrgb
|
estrogen-related receptor gamma b |
| chr12_-_10409961 | 0.95 |
ENSDART00000149521
ENSDART00000052001 |
eef2k
|
eukaryotic elongation factor 2 kinase |
| chr11_+_13176568 | 0.95 |
ENSDART00000125371
ENSDART00000123257 |
mknk1
|
MAP kinase interacting serine/threonine kinase 1 |
| chr24_-_41220538 | 0.93 |
ENSDART00000150207
|
acvr2ba
|
activin A receptor type 2Ba |
| chr15_+_46356879 | 0.92 |
ENSDART00000154388
|
wu:fb18f06
|
wu:fb18f06 |
| chr16_-_6944927 | 0.92 |
ENSDART00000149620
|
pmvk
|
phosphomevalonate kinase |
| chr5_-_25583125 | 0.90 |
ENSDART00000031665
ENSDART00000145353 |
anxa1a
|
annexin A1a |
| chr4_+_4509996 | 0.89 |
ENSDART00000028694
|
gnsa
|
glucosamine (N-acetyl)-6-sulfatase a |
| chr20_-_45812144 | 0.89 |
ENSDART00000147897
ENSDART00000147637 |
fermt1
|
fermitin family member 1 |
| chr5_-_25582721 | 0.88 |
ENSDART00000123986
|
anxa1a
|
annexin A1a |
| chr8_-_43158486 | 0.88 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr9_+_34319333 | 0.88 |
ENSDART00000035522
ENSDART00000146480 |
pou2f1b
|
POU class 2 homeobox 1b |
| chr3_+_45687266 | 0.86 |
ENSDART00000131652
|
gpr146
|
G protein-coupled receptor 146 |
| chr22_-_4439311 | 0.86 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr18_+_31016379 | 0.85 |
ENSDART00000172461
ENSDART00000163471 |
uraha
|
urate (5-hydroxyiso-) hydrolase a |
| chr3_-_46410387 | 0.85 |
ENSDART00000156822
|
cdip1
|
cell death-inducing p53 target 1 |
| chr17_+_42842632 | 0.84 |
ENSDART00000155547
ENSDART00000126087 |
qpct
|
glutaminyl-peptide cyclotransferase |
| chr10_-_35103208 | 0.84 |
ENSDART00000192734
|
zgc:110006
|
zgc:110006 |
| chr14_-_15155384 | 0.83 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr17_-_26926577 | 0.83 |
ENSDART00000050202
|
rcan3
|
regulator of calcineurin 3 |
| chr12_+_22560067 | 0.82 |
ENSDART00000172066
|
polr2a
|
polymerase (RNA) II (DNA directed) polypeptide A |
| chr18_-_15551360 | 0.81 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr15_-_31514818 | 0.81 |
ENSDART00000153978
|
hmgb1b
|
high mobility group box 1b |
| chr15_-_43284021 | 0.81 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| chr22_+_38301365 | 0.79 |
ENSDART00000137339
|
traf5
|
Tnf receptor-associated factor 5 |
| chr11_-_37997419 | 0.79 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr15_-_29354020 | 0.79 |
ENSDART00000127795
|
tsku
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis) |
| chr15_+_46357080 | 0.78 |
ENSDART00000155571
ENSDART00000156541 |
wu:fb18f06
|
wu:fb18f06 |
| chr1_+_51039558 | 0.78 |
ENSDART00000024743
|
dpy30
|
dpy-30 histone methyltransferase complex regulatory subunit |
| chr6_-_8264751 | 0.78 |
ENSDART00000091628
|
ccdc151
|
coiled-coil domain containing 151 |
| chr18_-_21950751 | 0.75 |
ENSDART00000043452
|
tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr17_-_25649079 | 0.73 |
ENSDART00000130955
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
| chr4_-_390431 | 0.73 |
ENSDART00000067482
ENSDART00000138500 |
dynlt1
|
dynein, light chain, Tctex-type 1 |
| chr5_-_65662996 | 0.72 |
ENSDART00000147707
|
camsap1b
|
calmodulin regulated spectrin-associated protein 1b |
| chr17_+_24109012 | 0.69 |
ENSDART00000156251
|
ehbp1
|
EH domain binding protein 1 |
| chr3_-_31716157 | 0.68 |
ENSDART00000193189
|
ccdc47
|
coiled-coil domain containing 47 |
| chr12_-_35582683 | 0.66 |
ENSDART00000167933
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr11_+_41858807 | 0.66 |
ENSDART00000161605
|
iffo2b
|
intermediate filament family orphan 2b |
| chr5_-_54481692 | 0.66 |
ENSDART00000165719
|
fbxw5
|
F-box and WD repeat domain containing 5 |
| chr10_-_31015535 | 0.65 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr6_+_6924637 | 0.65 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr6_-_6346557 | 0.65 |
ENSDART00000115018
|
ccdc88aa
|
coiled-coil domain containing 88Aa |
| chr2_-_42558549 | 0.65 |
ENSDART00000025997
|
dip2cb
|
disco-interacting protein 2 homolog Cb |
| chr17_-_11466700 | 0.65 |
ENSDART00000091159
|
adpgk2
|
ADP-dependent glucokinase 2 |
| chr20_-_3087162 | 0.64 |
ENSDART00000152495
|
map3k5
|
mitogen-activated protein kinase kinase kinase 5 |
| chr8_-_4431971 | 0.64 |
ENSDART00000141728
ENSDART00000133519 |
rad9b
|
RAD9 checkpoint clamp component B |
| chr20_-_5369105 | 0.63 |
ENSDART00000114316
|
sptlc2b
|
serine palmitoyltransferase, long chain base subunit 2b |
| chr5_-_13206878 | 0.63 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr4_+_6869847 | 0.63 |
ENSDART00000036646
|
dock4b
|
dedicator of cytokinesis 4b |
| chr18_+_38885309 | 0.63 |
ENSDART00000041597
|
arpp19a
|
cAMP-regulated phosphoprotein 19a |
| chr11_+_36409457 | 0.63 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr25_+_4787431 | 0.61 |
ENSDART00000170640
|
myo5c
|
myosin VC |
| chr20_-_1268863 | 0.61 |
ENSDART00000109321
ENSDART00000027119 |
lats1
|
large tumor suppressor kinase 1 |
| chr4_-_12795436 | 0.60 |
ENSDART00000131026
ENSDART00000075127 |
b2m
|
beta-2-microglobulin |
| chr20_+_2589414 | 0.60 |
ENSDART00000043626
|
il20ra
|
interleukin 20 receptor, alpha |
| chr12_-_33706726 | 0.59 |
ENSDART00000153135
|
myo15b
|
myosin XVB |
| chr2_-_23572104 | 0.59 |
ENSDART00000144083
|
si:dkey-58b18.6
|
si:dkey-58b18.6 |
| chr5_-_11809710 | 0.59 |
ENSDART00000186998
ENSDART00000181363 ENSDART00000180681 |
nf2a
|
neurofibromin 2a (merlin) |
| chr8_-_28274251 | 0.59 |
ENSDART00000050671
|
rap1aa
|
RAP1A, member of RAS oncogene family a |
| chr3_-_5067585 | 0.59 |
ENSDART00000169609
|
tefb
|
thyrotrophic embryonic factor b |
| chr15_-_47857687 | 0.58 |
ENSDART00000098982
ENSDART00000151594 |
h3f3b.1
|
H3 histone, family 3B.1 |
| chr20_+_18232893 | 0.58 |
ENSDART00000189225
|
kctd1
|
potassium channel tetramerization domain containing 1 |
| chr24_+_39186940 | 0.58 |
ENSDART00000155817
|
spsb3b
|
splA/ryanodine receptor domain and SOCS box containing 3b |
| chr25_+_19238175 | 0.58 |
ENSDART00000110730
ENSDART00000193619 ENSDART00000154420 |
ppip5k1b
|
diphosphoinositol pentakisphosphate kinase 1b |
| chr21_+_43328685 | 0.57 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr14_-_34512859 | 0.57 |
ENSDART00000140368
|
si:ch211-232m8.3
|
si:ch211-232m8.3 |
| chr3_-_30885250 | 0.57 |
ENSDART00000109104
|
kmt5c
|
lysine methyltransferase 5C |
| chr18_+_30567945 | 0.57 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr11_-_15001639 | 0.57 |
ENSDART00000170627
ENSDART00000168222 ENSDART00000171210 ENSDART00000163518 ENSDART00000168180 ENSDART00000157970 ENSDART00000169113 ENSDART00000180049 ENSDART00000188961 ENSDART00000183952 ENSDART00000193904 ENSDART00000193450 ENSDART00000186508 ENSDART00000169476 ENSDART00000162938 |
elavl1b
|
ELAV like RNA binding protein 1b |
| chr24_-_26981848 | 0.56 |
ENSDART00000183198
|
stag1b
|
stromal antigen 1b |
| chr23_-_25798099 | 0.55 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr5_-_22615087 | 0.54 |
ENSDART00000146035
|
zgc:113208
|
zgc:113208 |
| chr20_+_22045089 | 0.54 |
ENSDART00000063564
ENSDART00000187013 ENSDART00000161552 ENSDART00000174478 ENSDART00000063568 ENSDART00000152247 |
nmu
|
neuromedin U |
| chr1_-_36151377 | 0.54 |
ENSDART00000037516
|
znf827
|
zinc finger protein 827 |
| chr17_+_21964472 | 0.53 |
ENSDART00000063704
ENSDART00000188904 |
crip3
|
cysteine-rich protein 3 |
| chr2_-_55298075 | 0.53 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr9_-_7212973 | 0.53 |
ENSDART00000133638
|
mgat4a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr1_+_51496862 | 0.53 |
ENSDART00000150433
|
meis1a
|
Meis homeobox 1 a |
| chr5_+_44228526 | 0.53 |
ENSDART00000143789
|
TMEM8B
|
si:dkey-84j12.1 |
| chr17_-_2039511 | 0.52 |
ENSDART00000160223
|
spint1a
|
serine peptidase inhibitor, Kunitz type 1 a |
| chr2_+_6243144 | 0.52 |
ENSDART00000058258
|
gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr1_-_56080112 | 0.51 |
ENSDART00000075469
ENSDART00000161473 |
c3a.6
|
complement component c3a, duplicate 6 |
| chr20_-_1239596 | 0.51 |
ENSDART00000049063
ENSDART00000140650 |
ankrd6b
|
ankyrin repeat domain 6b |
| chr12_-_19282120 | 0.51 |
ENSDART00000153260
|
si:ch211-141o9.10
|
si:ch211-141o9.10 |
| chr18_+_5917625 | 0.50 |
ENSDART00000169100
|
glg1b
|
golgi glycoprotein 1b |
| chr24_+_39614853 | 0.50 |
ENSDART00000165138
|
BX547934.2
|
|
| chr21_-_26071773 | 0.50 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr13_+_47821524 | 0.50 |
ENSDART00000109978
|
zc3h6
|
zinc finger CCCH-type containing 6 |
| chr6_+_58289335 | 0.50 |
ENSDART00000177399
|
ralgapb
|
Ral GTPase activating protein, beta subunit (non-catalytic) |
| chr17_-_49438873 | 0.50 |
ENSDART00000004424
|
znf292a
|
zinc finger protein 292a |
| chr8_+_49433663 | 0.49 |
ENSDART00000140481
ENSDART00000180714 |
nfu1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr2_-_16159491 | 0.48 |
ENSDART00000110059
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr19_-_3106447 | 0.48 |
ENSDART00000137987
|
si:ch211-80h18.1
|
si:ch211-80h18.1 |
| chr14_-_4044545 | 0.47 |
ENSDART00000169527
|
snx25
|
sorting nexin 25 |
| chr14_+_14662116 | 0.47 |
ENSDART00000161693
|
cetn2
|
centrin, EF-hand protein, 2 |
| chr18_+_3579829 | 0.47 |
ENSDART00000158763
ENSDART00000182850 ENSDART00000162754 ENSDART00000178789 ENSDART00000172656 |
lrch3
|
leucine-rich repeats and calponin homology (CH) domain containing 3 |
| chr16_+_26706519 | 0.47 |
ENSDART00000142706
|
virma
|
vir like m6A methyltransferase associated |
| chr3_+_1107102 | 0.46 |
ENSDART00000092690
|
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr6_+_7322587 | 0.46 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr9_+_54179306 | 0.46 |
ENSDART00000189829
|
tmsb4x
|
thymosin, beta 4 x |
| chr11_-_45138857 | 0.45 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr22_-_347424 | 0.45 |
ENSDART00000067633
|
necap2
|
NECAP endocytosis associated 2 |
| chr8_-_8698607 | 0.45 |
ENSDART00000046712
|
zgc:86609
|
zgc:86609 |
| chr23_+_39606108 | 0.45 |
ENSDART00000109464
|
g0s2
|
G0/G1 switch 2 |
| chr2_+_19633493 | 0.44 |
ENSDART00000147989
|
pimr54
|
Pim proto-oncogene, serine/threonine kinase, related 54 |
| chr25_-_25058508 | 0.44 |
ENSDART00000087570
ENSDART00000178891 |
FQ311928.1
|
|
| chr9_+_38481780 | 0.42 |
ENSDART00000087241
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr13_+_34689663 | 0.42 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr24_+_792429 | 0.42 |
ENSDART00000082523
|
impa2
|
inositol(myo)-1(or 4)-monophosphatase 2 |
| chr20_+_29690901 | 0.42 |
ENSDART00000142669
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr19_+_935565 | 0.42 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr21_-_3613702 | 0.42 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr25_+_23280220 | 0.41 |
ENSDART00000153940
|
ptprjb.1
|
protein tyrosine phosphatase, receptor type, Jb, tandem duplicate 1 |
| chr21_-_25295087 | 0.41 |
ENSDART00000087910
ENSDART00000147860 |
st14b
|
suppression of tumorigenicity 14 (colon carcinoma) b |
| chr24_-_39772045 | 0.41 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr8_-_7603516 | 0.41 |
ENSDART00000179826
ENSDART00000190153 |
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr7_+_33424044 | 0.41 |
ENSDART00000180260
|
glceb
|
glucuronic acid epimerase b |
| chr1_-_7570181 | 0.41 |
ENSDART00000103588
|
mxa
|
myxovirus (influenza) resistance A |
| chr21_-_34032650 | 0.41 |
ENSDART00000138575
ENSDART00000047515 |
rnf145b
|
ring finger protein 145b |
| chr13_+_39236141 | 0.41 |
ENSDART00000111458
|
zgc:172136
|
zgc:172136 |
| chr16_-_4531657 | 0.41 |
ENSDART00000028419
|
ppp1r8b
|
protein phosphatase 1, regulatory subunit 8b |
| chr8_-_47152001 | 0.40 |
ENSDART00000163922
ENSDART00000110512 ENSDART00000024320 |
ybx1
|
Y box binding protein 1 |
| chr7_-_4110462 | 0.39 |
ENSDART00000173318
|
zgc:55733
|
zgc:55733 |
| chr15_+_36310442 | 0.39 |
ENSDART00000154678
|
gmnc
|
geminin coiled-coil domain containing |
| chr10_-_27566481 | 0.39 |
ENSDART00000078920
|
auts2a
|
autism susceptibility candidate 2a |
| chr10_+_18911152 | 0.39 |
ENSDART00000030205
|
bnip3lb
|
BCL2 interacting protein 3 like b |
| chr15_+_25024685 | 0.38 |
ENSDART00000188889
|
BX908796.7
|
|
| chr3_-_23407720 | 0.38 |
ENSDART00000155658
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr12_+_48803098 | 0.38 |
ENSDART00000074768
|
ppifb
|
peptidylprolyl isomerase Fb |
| chr21_-_45073764 | 0.38 |
ENSDART00000181390
ENSDART00000063714 |
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr4_+_12292274 | 0.38 |
ENSDART00000061070
ENSDART00000150786 |
mkrn1
|
makorin, ring finger protein, 1 |
| chr1_+_26445615 | 0.37 |
ENSDART00000180810
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr8_-_7603700 | 0.37 |
ENSDART00000137975
|
irak1
|
interleukin-1 receptor-associated kinase 1 |
| chr2_-_16159203 | 0.37 |
ENSDART00000153480
|
vav3b
|
vav 3 guanine nucleotide exchange factor b |
| chr3_+_24537023 | 0.37 |
ENSDART00000077702
|
sp100.1
|
SP110 nuclear body protein, tandem duplicate 1 |
| chr8_+_22931427 | 0.37 |
ENSDART00000063096
|
sypa
|
synaptophysin a |
| chr1_-_51157660 | 0.37 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr7_+_32369026 | 0.36 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr19_+_7424347 | 0.36 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr12_-_11593436 | 0.36 |
ENSDART00000138954
|
plekha1b
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1b |
| chr25_+_5012791 | 0.35 |
ENSDART00000156970
|
si:ch73-265h17.5
|
si:ch73-265h17.5 |
| chr7_-_44704910 | 0.35 |
ENSDART00000037850
|
dync1li2
|
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr20_+_29691118 | 0.35 |
ENSDART00000164121
|
mboat2b
|
membrane bound O-acyltransferase domain containing 2b |
| chr3_-_6519691 | 0.34 |
ENSDART00000165273
ENSDART00000179882 ENSDART00000172292 |
GGA3 (1 of many)
|
si:ch73-157i16.3 |
| chr14_-_33312407 | 0.34 |
ENSDART00000135700
|
sept6
|
septin 6 |
| chr16_-_31451720 | 0.34 |
ENSDART00000146886
|
csnk2a1
|
casein kinase 2, alpha 1 polypeptide |
| chr14_-_49133426 | 0.34 |
ENSDART00000042421
|
ppp2ca
|
protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr3_-_39305291 | 0.34 |
ENSDART00000102674
|
plcd3a
|
phospholipase C, delta 3a |
| chr4_-_17263210 | 0.34 |
ENSDART00000147853
|
lrmp
|
lymphoid-restricted membrane protein |
| chr9_-_32847642 | 0.34 |
ENSDART00000121506
|
hpx
|
hemopexin |
| chr4_-_2219705 | 0.34 |
ENSDART00000131046
|
si:ch73-278m9.1
|
si:ch73-278m9.1 |
| chr11_+_30162407 | 0.34 |
ENSDART00000190333
ENSDART00000127502 |
cdkl5
|
cyclin-dependent kinase-like 5 |
| chr24_+_5208171 | 0.34 |
ENSDART00000155926
ENSDART00000154464 |
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr5_+_45138934 | 0.33 |
ENSDART00000041412
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_8643901 | 0.33 |
ENSDART00000142076
ENSDART00000075624 |
usp11
|
ubiquitin specific peptidase 11 |
| chr5_+_44190974 | 0.33 |
ENSDART00000182634
ENSDART00000190626 |
TMEM8B
|
si:dkey-84j12.1 |
| chr5_+_6954162 | 0.33 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr18_+_20869923 | 0.32 |
ENSDART00000138471
|
pgpep1l
|
pyroglutamyl-peptidase I-like |
| chr13_-_2215213 | 0.32 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr14_-_4121052 | 0.32 |
ENSDART00000167074
|
irf2
|
interferon regulatory factor 2 |
| chr19_+_1510971 | 0.32 |
ENSDART00000157721
|
SLC45A4 (1 of many)
|
solute carrier family 45 member 4 |
| chr8_+_7097740 | 0.31 |
ENSDART00000159670
|
abtb1
|
ankyrin repeat and BTB (POZ) domain containing 1 |
| chr18_+_40617717 | 0.31 |
ENSDART00000049423
|
zgc:158427
|
zgc:158427 |
| chr13_-_34683370 | 0.31 |
ENSDART00000113661
|
kif16bb
|
kinesin family member 16Bb |
| chr5_+_46077710 | 0.31 |
ENSDART00000084069
ENSDART00000138055 |
iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr16_+_25074029 | 0.31 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr3_-_60175470 | 0.31 |
ENSDART00000156597
|
si:ch73-364h19.1
|
si:ch73-364h19.1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of pou2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.4 | 1.1 | GO:0071830 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.3 | 1.0 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 2.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.3 | 1.8 | GO:0032743 | neutrophil apoptotic process(GO:0001781) regulation of T-helper 1 type immune response(GO:0002825) positive regulation of T-helper 1 type immune response(GO:0002827) negative regulation of type 2 immune response(GO:0002829) inflammatory cell apoptotic process(GO:0006925) regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of phospholipase A2 activity(GO:0032429) interleukin-2 production(GO:0032623) regulation of interleukin-2 production(GO:0032663) positive regulation of interleukin-2 production(GO:0032743) myeloid cell apoptotic process(GO:0033028) regulation of neutrophil apoptotic process(GO:0033029) positive regulation of neutrophil apoptotic process(GO:0033031) regulation of myeloid cell apoptotic process(GO:0033032) positive regulation of myeloid cell apoptotic process(GO:0033034) T-helper 1 type immune response(GO:0042088) positive regulation of CD4-positive, alpha-beta T cell differentiation(GO:0043372) T-helper 1 cell differentiation(GO:0045063) positive regulation of T-helper cell differentiation(GO:0045624) regulation of T-helper 1 cell differentiation(GO:0045625) positive regulation of T-helper 1 cell differentiation(GO:0045627) regulation of T-helper 2 cell differentiation(GO:0045628) negative regulation of T-helper 2 cell differentiation(GO:0045629) positive regulation of fatty acid biosynthetic process(GO:0045723) positive regulation of alpha-beta T cell differentiation(GO:0046638) neutrophil clearance(GO:0097350) negative regulation of phospholipase A2 activity(GO:1900138) positive regulation of CD4-positive, alpha-beta T cell activation(GO:2000516) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.3 | 0.8 | GO:0046415 | urate catabolic process(GO:0019628) urate metabolic process(GO:0046415) |
| 0.3 | 2.0 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.3 | 1.1 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.2 | 1.2 | GO:1901906 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 0.9 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.2 | 1.1 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.2 | 1.3 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.2 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.2 | 0.8 | GO:0018199 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) peptidyl-glutamine modification(GO:0018199) |
| 0.2 | 0.8 | GO:0003418 | chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.2 | 0.6 | GO:0034770 | histone H4-K20 methylation(GO:0034770) histone H4-K20 trimethylation(GO:0034773) |
| 0.2 | 0.6 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 1.0 | GO:0032475 | otolith formation(GO:0032475) |
| 0.1 | 1.0 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 0.5 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.6 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.1 | 0.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.6 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:0035046 | pronuclear migration(GO:0035046) |
| 0.1 | 0.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.0 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.1 | 0.8 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.8 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.9 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.1 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.6 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 0.6 | GO:0006111 | regulation of gluconeogenesis(GO:0006111) |
| 0.1 | 0.4 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 | 1.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.4 | GO:1903523 | negative regulation of heart contraction(GO:0045822) negative regulation of blood circulation(GO:1903523) |
| 0.1 | 1.0 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 1.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 1.0 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.8 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.1 | 0.4 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.1 | 0.2 | GO:0046831 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.1 | 0.4 | GO:0046685 | response to arsenic-containing substance(GO:0046685) |
| 0.1 | 0.4 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.8 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.1 | 0.3 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 0.5 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.1 | 0.6 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.1 | 0.3 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 1.0 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 0.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.6 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.3 | GO:0046532 | amacrine cell differentiation(GO:0035881) regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.9 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.5 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.4 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.5 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.7 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.5 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.0 | 0.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.8 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.9 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.4 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.7 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.0 | 0.2 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.3 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.5 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.2 | GO:0038065 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 0.3 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 2.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine metabolic process(GO:0046102) inosine biosynthetic process(GO:0046103) |
| 0.0 | 1.1 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.0 | 0.2 | GO:0006264 | mitochondrial DNA replication(GO:0006264) mitochondrial DNA metabolic process(GO:0032042) |
| 0.0 | 0.1 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.1 | GO:0035666 | regulation of natural killer cell activation(GO:0032814) positive regulation of natural killer cell activation(GO:0032816) TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 0.0 | GO:0051589 | negative regulation of neurotransmitter transport(GO:0051589) |
| 0.0 | 0.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.1 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.0 | 0.2 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.5 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.1 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.4 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.0 | 0.5 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.2 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.5 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.0 | 0.9 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.5 | GO:0048264 | determination of ventral identity(GO:0048264) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.2 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.1 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.0 | 1.9 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0045109 | intermediate filament organization(GO:0045109) intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.4 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0035066 | positive regulation of histone acetylation(GO:0035066) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0061621 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.6 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.6 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
| 0.0 | 0.0 | GO:0033212 | iron assimilation(GO:0033212) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0072015 | glomerular filtration(GO:0003094) glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.3 | 1.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.2 | 0.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 2.5 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 0.6 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 1.8 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.1 | 1.0 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.4 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 0.9 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.6 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 0.9 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.6 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.8 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 1.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0070209 | ASTRA complex(GO:0070209) |
| 0.0 | 1.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:0098645 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.7 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 2.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.6 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.2 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0098844 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.5 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.1 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.2 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.4 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.4 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.4 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.3 | 1.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.3 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.2 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 0.8 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.2 | 0.6 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.2 | 2.0 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.2 | 0.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 1.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 2.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.6 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.4 | GO:0052833 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.1 | 0.8 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.1 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.4 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.1 | 0.4 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.1 | 0.6 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.1 | 2.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 1.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) |
| 0.1 | 0.4 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.1 | 1.0 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 1.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 0.9 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.1 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.2 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.8 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.4 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 1.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.6 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 1.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.0 | 1.0 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.0 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.5 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.5 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.6 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.2 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 0.0 | 0.4 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 3.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.6 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.1 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.9 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.1 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.5 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004408 | holocytochrome-c synthase activity(GO:0004408) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 1.4 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.7 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.1 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.0 | 3.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.1 | 1.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.6 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 2.4 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.2 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.6 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.9 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.9 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 0.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 2.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 1.0 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.3 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.8 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.0 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.0 | 1.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |